
Primorskyibacter sp. SS33
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Primorskyibacter; unclassified Primorskyibacter
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
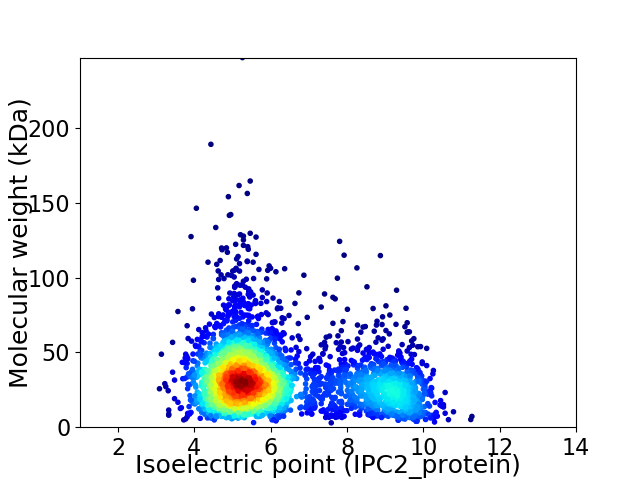
Virtual 2D-PAGE plot for 3259 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6APE9|A0A4R6APE9_9RHOB Glycosyltransferase OS=Primorskyibacter sp. SS33 OX=2547833 GN=E2L08_02385 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 8.88PTMFTTTAGLALCLGAGAALAQEE25 pKa = 4.31VTLTVEE31 pKa = 4.12SWRR34 pKa = 11.84SEE36 pKa = 3.95DD37 pKa = 3.35QAIWNDD43 pKa = 3.49TLIPAFEE50 pKa = 4.52EE51 pKa = 3.77KK52 pKa = 10.66HH53 pKa = 5.73PDD55 pKa = 2.93INIEE59 pKa = 4.11FTPTAPTEE67 pKa = 3.95YY68 pKa = 10.48NAALNARR75 pKa = 11.84LEE77 pKa = 4.43GGTAGDD83 pKa = 4.3LVTCRR88 pKa = 11.84SFDD91 pKa = 3.26GSLPLFNKK99 pKa = 9.94GQLADD104 pKa = 3.82ISDD107 pKa = 4.59LDD109 pKa = 3.97GLSNFPDD116 pKa = 4.73FARR119 pKa = 11.84AAWSTDD125 pKa = 2.91DD126 pKa = 4.13GASTFCVPMASVIHH140 pKa = 5.47GFIYY144 pKa = 10.7NADD147 pKa = 3.44AFAEE151 pKa = 4.38LGLEE155 pKa = 4.09EE156 pKa = 5.35PEE158 pKa = 4.31TVDD161 pKa = 4.01EE162 pKa = 5.24FFAVLDD168 pKa = 4.77AIKK171 pKa = 10.44EE172 pKa = 4.13DD173 pKa = 3.69GSYY176 pKa = 10.6IPLAMGSADD185 pKa = 3.15QWEE188 pKa = 4.8AATMAYY194 pKa = 9.69TNIGPNYY201 pKa = 8.24WKK203 pKa = 10.85GEE205 pKa = 4.12EE206 pKa = 4.34GRR208 pKa = 11.84QALIDD213 pKa = 3.53GTAAFTDD220 pKa = 4.01PQFVAPYY227 pKa = 8.69EE228 pKa = 4.1QMVRR232 pKa = 11.84WTDD235 pKa = 3.42YY236 pKa = 10.29MGPAYY241 pKa = 9.74QAQGYY246 pKa = 9.82SDD248 pKa = 3.84SQNLFTLGRR257 pKa = 11.84AAIYY261 pKa = 8.44PAGSWEE267 pKa = 3.81ITGFTEE273 pKa = 4.83AADD276 pKa = 4.02FEE278 pKa = 4.89MGAFPPPVQNAGDD291 pKa = 3.38TCYY294 pKa = 10.68ISDD297 pKa = 4.01HH298 pKa = 6.0TDD300 pKa = 2.37IGLGMNAATDD310 pKa = 3.65HH311 pKa = 6.98PEE313 pKa = 3.65AARR316 pKa = 11.84TFLEE320 pKa = 4.7WVASPEE326 pKa = 3.92FAEE329 pKa = 5.43LYY331 pKa = 10.16TNALPGFFSLSDD343 pKa = 3.36AQFEE347 pKa = 4.49VDD349 pKa = 4.6NPLAQEE355 pKa = 4.43FIEE358 pKa = 4.36FRR360 pKa = 11.84EE361 pKa = 4.25NCEE364 pKa = 3.73STVRR368 pKa = 11.84VASQILSRR376 pKa = 11.84GEE378 pKa = 3.78PNTWNNMWVLSANVLNDD395 pKa = 3.09AATPEE400 pKa = 4.15EE401 pKa = 4.38ATAEE405 pKa = 4.11LQQGLASWYY414 pKa = 9.83EE415 pKa = 3.92PQQNN419 pKa = 3.13
MM1 pKa = 7.55KK2 pKa = 8.88PTMFTTTAGLALCLGAGAALAQEE25 pKa = 4.31VTLTVEE31 pKa = 4.12SWRR34 pKa = 11.84SEE36 pKa = 3.95DD37 pKa = 3.35QAIWNDD43 pKa = 3.49TLIPAFEE50 pKa = 4.52EE51 pKa = 3.77KK52 pKa = 10.66HH53 pKa = 5.73PDD55 pKa = 2.93INIEE59 pKa = 4.11FTPTAPTEE67 pKa = 3.95YY68 pKa = 10.48NAALNARR75 pKa = 11.84LEE77 pKa = 4.43GGTAGDD83 pKa = 4.3LVTCRR88 pKa = 11.84SFDD91 pKa = 3.26GSLPLFNKK99 pKa = 9.94GQLADD104 pKa = 3.82ISDD107 pKa = 4.59LDD109 pKa = 3.97GLSNFPDD116 pKa = 4.73FARR119 pKa = 11.84AAWSTDD125 pKa = 2.91DD126 pKa = 4.13GASTFCVPMASVIHH140 pKa = 5.47GFIYY144 pKa = 10.7NADD147 pKa = 3.44AFAEE151 pKa = 4.38LGLEE155 pKa = 4.09EE156 pKa = 5.35PEE158 pKa = 4.31TVDD161 pKa = 4.01EE162 pKa = 5.24FFAVLDD168 pKa = 4.77AIKK171 pKa = 10.44EE172 pKa = 4.13DD173 pKa = 3.69GSYY176 pKa = 10.6IPLAMGSADD185 pKa = 3.15QWEE188 pKa = 4.8AATMAYY194 pKa = 9.69TNIGPNYY201 pKa = 8.24WKK203 pKa = 10.85GEE205 pKa = 4.12EE206 pKa = 4.34GRR208 pKa = 11.84QALIDD213 pKa = 3.53GTAAFTDD220 pKa = 4.01PQFVAPYY227 pKa = 8.69EE228 pKa = 4.1QMVRR232 pKa = 11.84WTDD235 pKa = 3.42YY236 pKa = 10.29MGPAYY241 pKa = 9.74QAQGYY246 pKa = 9.82SDD248 pKa = 3.84SQNLFTLGRR257 pKa = 11.84AAIYY261 pKa = 8.44PAGSWEE267 pKa = 3.81ITGFTEE273 pKa = 4.83AADD276 pKa = 4.02FEE278 pKa = 4.89MGAFPPPVQNAGDD291 pKa = 3.38TCYY294 pKa = 10.68ISDD297 pKa = 4.01HH298 pKa = 6.0TDD300 pKa = 2.37IGLGMNAATDD310 pKa = 3.65HH311 pKa = 6.98PEE313 pKa = 3.65AARR316 pKa = 11.84TFLEE320 pKa = 4.7WVASPEE326 pKa = 3.92FAEE329 pKa = 5.43LYY331 pKa = 10.16TNALPGFFSLSDD343 pKa = 3.36AQFEE347 pKa = 4.49VDD349 pKa = 4.6NPLAQEE355 pKa = 4.43FIEE358 pKa = 4.36FRR360 pKa = 11.84EE361 pKa = 4.25NCEE364 pKa = 3.73STVRR368 pKa = 11.84VASQILSRR376 pKa = 11.84GEE378 pKa = 3.78PNTWNNMWVLSANVLNDD395 pKa = 3.09AATPEE400 pKa = 4.15EE401 pKa = 4.38ATAEE405 pKa = 4.11LQQGLASWYY414 pKa = 9.83EE415 pKa = 3.92PQQNN419 pKa = 3.13
Molecular weight: 45.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6AL34|A0A4R6AL34_9RHOB LysR family transcriptional regulator OS=Primorskyibacter sp. SS33 OX=2547833 GN=E2L08_01260 PE=3 SV=1
MM1 pKa = 7.54NLGQIGTMAVRR12 pKa = 11.84MILRR16 pKa = 11.84RR17 pKa = 11.84LMSRR21 pKa = 11.84GMNAGINRR29 pKa = 11.84AFGAARR35 pKa = 11.84TPGAAKK41 pKa = 9.91QGRR44 pKa = 11.84AAQVSGKK51 pKa = 7.85RR52 pKa = 11.84TKK54 pKa = 10.23QAMRR58 pKa = 11.84VARR61 pKa = 11.84RR62 pKa = 11.84FGRR65 pKa = 11.84FF66 pKa = 2.87
MM1 pKa = 7.54NLGQIGTMAVRR12 pKa = 11.84MILRR16 pKa = 11.84RR17 pKa = 11.84LMSRR21 pKa = 11.84GMNAGINRR29 pKa = 11.84AFGAARR35 pKa = 11.84TPGAAKK41 pKa = 9.91QGRR44 pKa = 11.84AAQVSGKK51 pKa = 7.85RR52 pKa = 11.84TKK54 pKa = 10.23QAMRR58 pKa = 11.84VARR61 pKa = 11.84RR62 pKa = 11.84FGRR65 pKa = 11.84FF66 pKa = 2.87
Molecular weight: 7.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1031760 |
26 |
2364 |
316.6 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.779 ± 0.067 | 0.867 ± 0.013 |
6.4 ± 0.042 | 5.835 ± 0.044 |
3.52 ± 0.023 | 9.227 ± 0.04 |
1.962 ± 0.022 | 4.868 ± 0.029 |
2.215 ± 0.033 | 10.035 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.565 ± 0.019 | 2.072 ± 0.021 |
5.54 ± 0.036 | 2.623 ± 0.022 |
8.077 ± 0.047 | 4.552 ± 0.025 |
5.245 ± 0.027 | 7.22 ± 0.037 |
1.443 ± 0.02 | 1.952 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
