
Steccherinum ochraceum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Steccherinaceae; Steccherinum
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
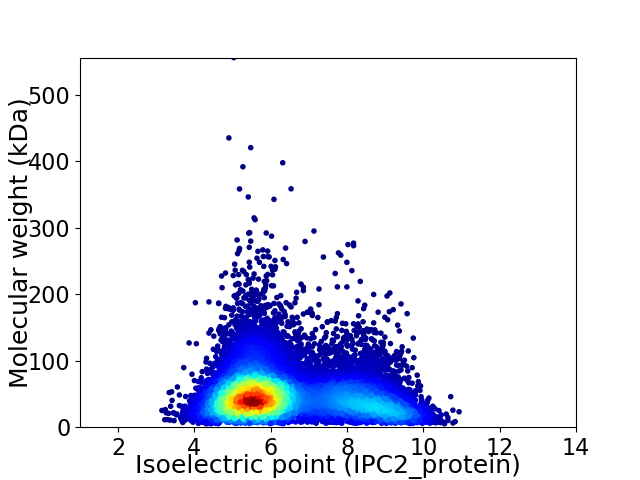
Virtual 2D-PAGE plot for 12246 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R0RHA8|A0A4R0RHA8_9APHY Uncharacterized protein OS=Steccherinum ochraceum OX=92696 GN=EIP91_004451 PE=3 SV=1
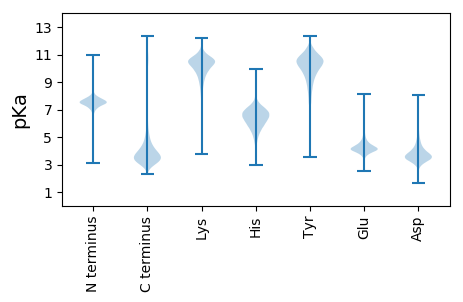
MM1 pKa = 7.44SLLSYY6 pKa = 10.11TLLALSLAGARR17 pKa = 11.84AATVSGSADD26 pKa = 3.28VNATAGASGSSVDD39 pKa = 3.7MVSAAWYY46 pKa = 7.66TGWHH50 pKa = 6.38SDD52 pKa = 2.97NFTLANVSWDD62 pKa = 3.42KK63 pKa = 9.27YY64 pKa = 7.17THH66 pKa = 5.01MTYY69 pKa = 10.72SFAITTPDD77 pKa = 3.24VKK79 pKa = 10.61QVSLDD84 pKa = 3.42GSDD87 pKa = 4.5PDD89 pKa = 4.56LLPQFVEE96 pKa = 4.27QAQKK100 pKa = 11.14NGVKK104 pKa = 10.47ALVSVGGWTGSLYY117 pKa = 10.67YY118 pKa = 10.42SSNVGSADD126 pKa = 3.03NRR128 pKa = 11.84TAFVKK133 pKa = 10.16TITDD137 pKa = 4.23FATQYY142 pKa = 11.05KK143 pKa = 10.42LDD145 pKa = 5.11GIDD148 pKa = 5.23FDD150 pKa = 4.0WEE152 pKa = 4.41YY153 pKa = 10.69PGTQGIGCNAISPDD167 pKa = 3.49DD168 pKa = 3.7TTNFLSFLQEE178 pKa = 3.85LRR180 pKa = 11.84QDD182 pKa = 3.69PVGKK186 pKa = 10.06NLFLTAATSITPWKK200 pKa = 10.23GPDD203 pKa = 3.35GTSLSDD209 pKa = 3.1VSGFAKK215 pKa = 10.49VLDD218 pKa = 4.58YY219 pKa = 10.84IAIMNYY225 pKa = 9.93DD226 pKa = 3.38INGAWSTAVGANAPLNDD243 pKa = 3.53TCAPAANQAGSAVSAVAAWTVAGMPSHH270 pKa = 6.36QIVLGVPSYY279 pKa = 10.98GHH281 pKa = 6.08SFHH284 pKa = 8.08VDD286 pKa = 2.83VEE288 pKa = 4.62DD289 pKa = 5.94AFHH292 pKa = 7.27CDD294 pKa = 5.4DD295 pKa = 4.72NGDD298 pKa = 4.05DD299 pKa = 3.99NGVLASFPPFDD310 pKa = 4.88KK311 pKa = 10.93AQQPSGDD318 pKa = 3.52AWDD321 pKa = 5.32DD322 pKa = 3.69GAGTDD327 pKa = 3.07ICGNQVAAGGNFDD340 pKa = 3.47FWGLIDD346 pKa = 5.41GGFLNANGTVADD358 pKa = 4.36GVFYY362 pKa = 10.53RR363 pKa = 11.84YY364 pKa = 9.79DD365 pKa = 3.44EE366 pKa = 4.8CSQTPYY372 pKa = 10.85VYY374 pKa = 10.91NEE376 pKa = 4.23DD377 pKa = 3.84SQVMVAFDD385 pKa = 4.29DD386 pKa = 4.07ATSFAAKK393 pKa = 10.22GDD395 pKa = 3.84FVKK398 pKa = 10.29TSNLRR403 pKa = 11.84GFAMWEE409 pKa = 3.82AGGDD413 pKa = 3.91SNDD416 pKa = 3.57ILLDD420 pKa = 3.9SIRR423 pKa = 11.84QAAGFEE429 pKa = 4.31TEE431 pKa = 6.04DD432 pKa = 5.14DD433 pKa = 4.84GEE435 pKa = 4.38DD436 pKa = 3.56CC437 pKa = 5.9
MM1 pKa = 7.44SLLSYY6 pKa = 10.11TLLALSLAGARR17 pKa = 11.84AATVSGSADD26 pKa = 3.28VNATAGASGSSVDD39 pKa = 3.7MVSAAWYY46 pKa = 7.66TGWHH50 pKa = 6.38SDD52 pKa = 2.97NFTLANVSWDD62 pKa = 3.42KK63 pKa = 9.27YY64 pKa = 7.17THH66 pKa = 5.01MTYY69 pKa = 10.72SFAITTPDD77 pKa = 3.24VKK79 pKa = 10.61QVSLDD84 pKa = 3.42GSDD87 pKa = 4.5PDD89 pKa = 4.56LLPQFVEE96 pKa = 4.27QAQKK100 pKa = 11.14NGVKK104 pKa = 10.47ALVSVGGWTGSLYY117 pKa = 10.67YY118 pKa = 10.42SSNVGSADD126 pKa = 3.03NRR128 pKa = 11.84TAFVKK133 pKa = 10.16TITDD137 pKa = 4.23FATQYY142 pKa = 11.05KK143 pKa = 10.42LDD145 pKa = 5.11GIDD148 pKa = 5.23FDD150 pKa = 4.0WEE152 pKa = 4.41YY153 pKa = 10.69PGTQGIGCNAISPDD167 pKa = 3.49DD168 pKa = 3.7TTNFLSFLQEE178 pKa = 3.85LRR180 pKa = 11.84QDD182 pKa = 3.69PVGKK186 pKa = 10.06NLFLTAATSITPWKK200 pKa = 10.23GPDD203 pKa = 3.35GTSLSDD209 pKa = 3.1VSGFAKK215 pKa = 10.49VLDD218 pKa = 4.58YY219 pKa = 10.84IAIMNYY225 pKa = 9.93DD226 pKa = 3.38INGAWSTAVGANAPLNDD243 pKa = 3.53TCAPAANQAGSAVSAVAAWTVAGMPSHH270 pKa = 6.36QIVLGVPSYY279 pKa = 10.98GHH281 pKa = 6.08SFHH284 pKa = 8.08VDD286 pKa = 2.83VEE288 pKa = 4.62DD289 pKa = 5.94AFHH292 pKa = 7.27CDD294 pKa = 5.4DD295 pKa = 4.72NGDD298 pKa = 4.05DD299 pKa = 3.99NGVLASFPPFDD310 pKa = 4.88KK311 pKa = 10.93AQQPSGDD318 pKa = 3.52AWDD321 pKa = 5.32DD322 pKa = 3.69GAGTDD327 pKa = 3.07ICGNQVAAGGNFDD340 pKa = 3.47FWGLIDD346 pKa = 5.41GGFLNANGTVADD358 pKa = 4.36GVFYY362 pKa = 10.53RR363 pKa = 11.84YY364 pKa = 9.79DD365 pKa = 3.44EE366 pKa = 4.8CSQTPYY372 pKa = 10.85VYY374 pKa = 10.91NEE376 pKa = 4.23DD377 pKa = 3.84SQVMVAFDD385 pKa = 4.29DD386 pKa = 4.07ATSFAAKK393 pKa = 10.22GDD395 pKa = 3.84FVKK398 pKa = 10.29TSNLRR403 pKa = 11.84GFAMWEE409 pKa = 3.82AGGDD413 pKa = 3.91SNDD416 pKa = 3.57ILLDD420 pKa = 3.9SIRR423 pKa = 11.84QAAGFEE429 pKa = 4.31TEE431 pKa = 6.04DD432 pKa = 5.14DD433 pKa = 4.84GEE435 pKa = 4.38DD436 pKa = 3.56CC437 pKa = 5.9
Molecular weight: 46.18 kDa
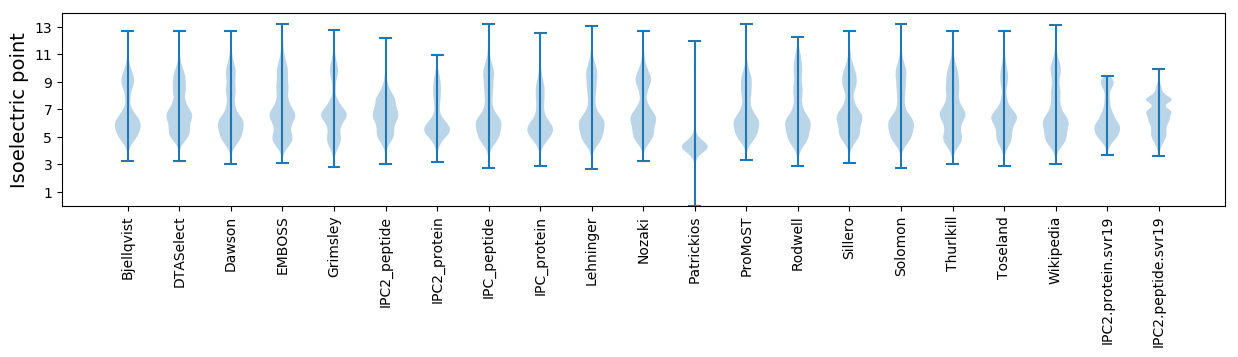
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R0RFQ8|A0A4R0RFQ8_9APHY F-box domain-containing protein OS=Steccherinum ochraceum OX=92696 GN=EIP91_006794 PE=4 SV=1
SS1 pKa = 7.04PLPHH5 pKa = 6.93RR6 pKa = 11.84RR7 pKa = 11.84LLLDD11 pKa = 3.71LRR13 pKa = 11.84CGSGRR18 pKa = 11.84AMILRR23 pKa = 11.84TMTGKK28 pKa = 9.82VLKK31 pKa = 8.04TTTTMLPPICSNARR45 pKa = 11.84TPHH48 pKa = 6.67PPPPPPHH55 pKa = 6.93RR56 pKa = 11.84PLPPQMRR63 pKa = 11.84PHH65 pKa = 6.59SRR67 pKa = 11.84QLPLNLGGTSGRR79 pKa = 11.84ATTRR83 pKa = 11.84ARR85 pKa = 11.84TQMSKK90 pKa = 10.3VVMIAKK96 pKa = 6.51TTLRR100 pKa = 11.84TSPTTLPNARR110 pKa = 11.84TPHH113 pKa = 6.72PPPPPPHH120 pKa = 7.0RR121 pKa = 11.84LLPPQMRR128 pKa = 11.84PHH130 pKa = 6.5SRR132 pKa = 11.84QLPINLGGISGRR144 pKa = 11.84AMTRR148 pKa = 11.84SRR150 pKa = 11.84AMAKK154 pKa = 8.72TVVMMLTVVRR164 pKa = 11.84TTAWSDD170 pKa = 3.34ARR172 pKa = 11.84PQSLSQQLPRR182 pKa = 11.84RR183 pKa = 11.84ALGGRR188 pKa = 11.84FRR190 pKa = 11.84GAMRR194 pKa = 11.84LMTRR198 pKa = 11.84ARR200 pKa = 11.84MVMMARR206 pKa = 11.84TFHH209 pKa = 6.41
SS1 pKa = 7.04PLPHH5 pKa = 6.93RR6 pKa = 11.84RR7 pKa = 11.84LLLDD11 pKa = 3.71LRR13 pKa = 11.84CGSGRR18 pKa = 11.84AMILRR23 pKa = 11.84TMTGKK28 pKa = 9.82VLKK31 pKa = 8.04TTTTMLPPICSNARR45 pKa = 11.84TPHH48 pKa = 6.67PPPPPPHH55 pKa = 6.93RR56 pKa = 11.84PLPPQMRR63 pKa = 11.84PHH65 pKa = 6.59SRR67 pKa = 11.84QLPLNLGGTSGRR79 pKa = 11.84ATTRR83 pKa = 11.84ARR85 pKa = 11.84TQMSKK90 pKa = 10.3VVMIAKK96 pKa = 6.51TTLRR100 pKa = 11.84TSPTTLPNARR110 pKa = 11.84TPHH113 pKa = 6.72PPPPPPHH120 pKa = 7.0RR121 pKa = 11.84LLPPQMRR128 pKa = 11.84PHH130 pKa = 6.5SRR132 pKa = 11.84QLPINLGGISGRR144 pKa = 11.84AMTRR148 pKa = 11.84SRR150 pKa = 11.84AMAKK154 pKa = 8.72TVVMMLTVVRR164 pKa = 11.84TTAWSDD170 pKa = 3.34ARR172 pKa = 11.84PQSLSQQLPRR182 pKa = 11.84RR183 pKa = 11.84ALGGRR188 pKa = 11.84FRR190 pKa = 11.84GAMRR194 pKa = 11.84LMTRR198 pKa = 11.84ARR200 pKa = 11.84MVMMARR206 pKa = 11.84TFHH209 pKa = 6.41
Molecular weight: 23.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5899370 |
50 |
5007 |
481.7 |
53.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.861 ± 0.018 | 1.131 ± 0.007 |
5.777 ± 0.015 | 5.858 ± 0.019 |
3.721 ± 0.014 | 6.346 ± 0.022 |
2.59 ± 0.01 | 4.602 ± 0.016 |
4.396 ± 0.018 | 9.268 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.095 ± 0.008 | 3.227 ± 0.012 |
6.75 ± 0.028 | 3.743 ± 0.012 |
6.149 ± 0.016 | 8.782 ± 0.027 |
6.093 ± 0.013 | 6.577 ± 0.015 |
1.416 ± 0.007 | 2.616 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
