
Brevibacterium luteolum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Brevibacteriaceae; Brevibacterium
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
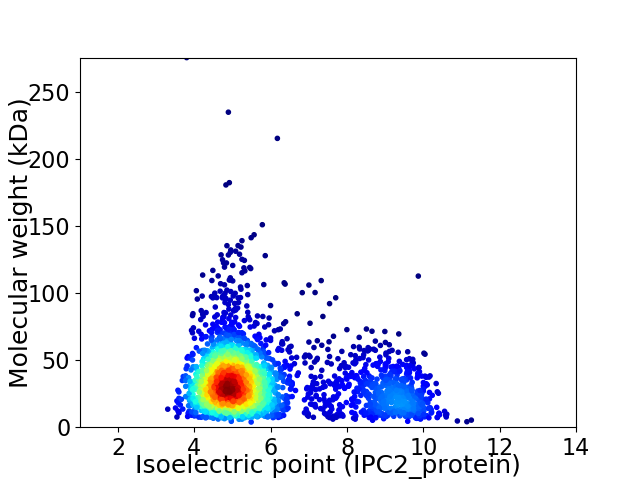
Virtual 2D-PAGE plot for 2715 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
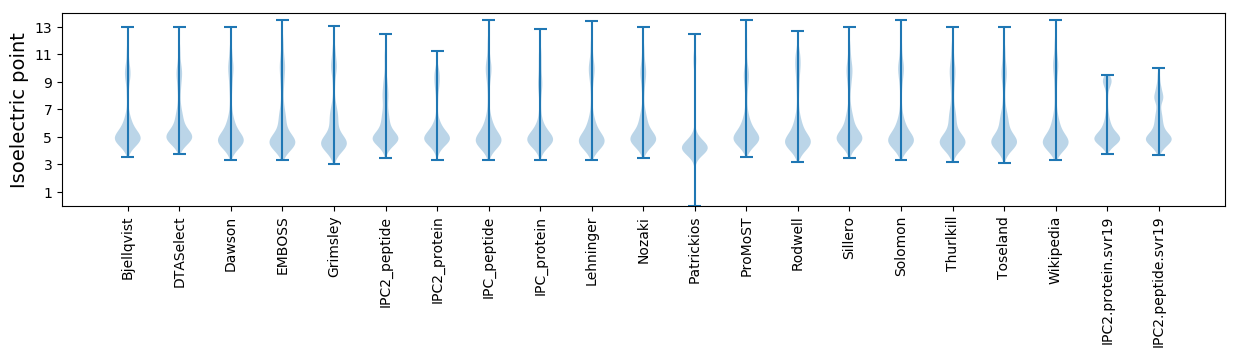
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N6PIE3|A0A2N6PIE3_9MICO Class I SAM-dependent methyltransferase OS=Brevibacterium luteolum OX=199591 GN=CJ198_03665 PE=4 SV=1
MM1 pKa = 7.56HH2 pKa = 7.53PFSHH6 pKa = 7.35PALALIAAGAALTLAAGCTAAEE28 pKa = 4.49DD29 pKa = 4.47APGSSPAPSTPEE41 pKa = 3.77PPQLSEE47 pKa = 5.57AITLDD52 pKa = 4.48FYY54 pKa = 11.97ASLLADD60 pKa = 3.82NDD62 pKa = 3.41TDD64 pKa = 3.75RR65 pKa = 11.84ALVADD70 pKa = 4.06AMGGRR75 pKa = 11.84LVAVGADD82 pKa = 3.75GAEE85 pKa = 4.29LWTHH89 pKa = 5.37QAQVDD94 pKa = 3.74MDD96 pKa = 4.24RR97 pKa = 11.84PDD99 pKa = 3.33SVAAYY104 pKa = 9.91SHH106 pKa = 6.88GEE108 pKa = 4.05TVIVDD113 pKa = 3.79DD114 pKa = 3.78GTGRR118 pKa = 11.84LRR120 pKa = 11.84ALDD123 pKa = 3.41WGDD126 pKa = 3.75GSEE129 pKa = 3.86AWTYY133 pKa = 10.29TPADD137 pKa = 3.76ADD139 pKa = 4.3LGCTEE144 pKa = 4.64PGDD147 pKa = 4.34YY148 pKa = 10.5YY149 pKa = 11.25SSSSTGTSALGEE161 pKa = 4.27DD162 pKa = 4.72GIILLEE168 pKa = 4.06YY169 pKa = 11.16GMIDD173 pKa = 3.75ADD175 pKa = 4.33RR176 pKa = 11.84GSCTGDD182 pKa = 3.12EE183 pKa = 4.25QKK185 pKa = 11.74AMVLGLDD192 pKa = 3.91PASGQPAWPAVQATADD208 pKa = 3.77AQPMGGANLQITPDD222 pKa = 3.41RR223 pKa = 11.84THH225 pKa = 7.39AYY227 pKa = 10.15LPWRR231 pKa = 11.84DD232 pKa = 3.16GDD234 pKa = 3.98GSFLTRR240 pKa = 11.84LDD242 pKa = 3.85LASGQHH248 pKa = 5.1ATVDD252 pKa = 3.36LSEE255 pKa = 4.8YY256 pKa = 10.92GEE258 pKa = 3.94QDD260 pKa = 3.3LGFGIYY266 pKa = 8.45PQKK269 pKa = 11.16APGTLVYY276 pKa = 10.89AFGTADD282 pKa = 3.55PDD284 pKa = 3.53APYY287 pKa = 10.29SGSVTRR293 pKa = 11.84TALLRR298 pKa = 11.84LPMDD302 pKa = 4.14LPEE305 pKa = 5.04SSEE308 pKa = 3.98ALTPLSAADD317 pKa = 4.44DD318 pKa = 4.46PGLAAGIDD326 pKa = 4.15FSEE329 pKa = 4.9EE330 pKa = 3.83PVDD333 pKa = 3.63YY334 pKa = 10.84CAARR338 pKa = 11.84PVSTASGTFVCASTQLFASRR358 pKa = 11.84ILVLGSLLDD367 pKa = 3.72GEE369 pKa = 5.07AWHH372 pKa = 6.82TDD374 pKa = 2.5ISEE377 pKa = 4.12DD378 pKa = 3.36AFEE381 pKa = 4.95FGLFNEE387 pKa = 4.76QYY389 pKa = 10.88GRR391 pKa = 11.84LHH393 pKa = 6.19TPVDD397 pKa = 3.74GTDD400 pKa = 3.32APLVIMPGLDD410 pKa = 2.8AGVRR414 pKa = 11.84AVSAVDD420 pKa = 5.48GSTLWEE426 pKa = 4.49AGEE429 pKa = 4.87DD430 pKa = 4.12LDD432 pKa = 5.44PLDD435 pKa = 4.7PVEE438 pKa = 4.24GSYY441 pKa = 11.58GFGGNGAFPQLGLIPVLLNDD461 pKa = 3.64AVHH464 pKa = 7.02FFTLDD469 pKa = 3.44GKK471 pKa = 9.95EE472 pKa = 3.98HH473 pKa = 6.86AEE475 pKa = 3.76AHH477 pKa = 6.22AVGQYY482 pKa = 10.7AEE484 pKa = 4.54PSATGRR490 pKa = 11.84WFAAAAGEE498 pKa = 4.52STTLWTVTGGG508 pKa = 3.29
MM1 pKa = 7.56HH2 pKa = 7.53PFSHH6 pKa = 7.35PALALIAAGAALTLAAGCTAAEE28 pKa = 4.49DD29 pKa = 4.47APGSSPAPSTPEE41 pKa = 3.77PPQLSEE47 pKa = 5.57AITLDD52 pKa = 4.48FYY54 pKa = 11.97ASLLADD60 pKa = 3.82NDD62 pKa = 3.41TDD64 pKa = 3.75RR65 pKa = 11.84ALVADD70 pKa = 4.06AMGGRR75 pKa = 11.84LVAVGADD82 pKa = 3.75GAEE85 pKa = 4.29LWTHH89 pKa = 5.37QAQVDD94 pKa = 3.74MDD96 pKa = 4.24RR97 pKa = 11.84PDD99 pKa = 3.33SVAAYY104 pKa = 9.91SHH106 pKa = 6.88GEE108 pKa = 4.05TVIVDD113 pKa = 3.79DD114 pKa = 3.78GTGRR118 pKa = 11.84LRR120 pKa = 11.84ALDD123 pKa = 3.41WGDD126 pKa = 3.75GSEE129 pKa = 3.86AWTYY133 pKa = 10.29TPADD137 pKa = 3.76ADD139 pKa = 4.3LGCTEE144 pKa = 4.64PGDD147 pKa = 4.34YY148 pKa = 10.5YY149 pKa = 11.25SSSSTGTSALGEE161 pKa = 4.27DD162 pKa = 4.72GIILLEE168 pKa = 4.06YY169 pKa = 11.16GMIDD173 pKa = 3.75ADD175 pKa = 4.33RR176 pKa = 11.84GSCTGDD182 pKa = 3.12EE183 pKa = 4.25QKK185 pKa = 11.74AMVLGLDD192 pKa = 3.91PASGQPAWPAVQATADD208 pKa = 3.77AQPMGGANLQITPDD222 pKa = 3.41RR223 pKa = 11.84THH225 pKa = 7.39AYY227 pKa = 10.15LPWRR231 pKa = 11.84DD232 pKa = 3.16GDD234 pKa = 3.98GSFLTRR240 pKa = 11.84LDD242 pKa = 3.85LASGQHH248 pKa = 5.1ATVDD252 pKa = 3.36LSEE255 pKa = 4.8YY256 pKa = 10.92GEE258 pKa = 3.94QDD260 pKa = 3.3LGFGIYY266 pKa = 8.45PQKK269 pKa = 11.16APGTLVYY276 pKa = 10.89AFGTADD282 pKa = 3.55PDD284 pKa = 3.53APYY287 pKa = 10.29SGSVTRR293 pKa = 11.84TALLRR298 pKa = 11.84LPMDD302 pKa = 4.14LPEE305 pKa = 5.04SSEE308 pKa = 3.98ALTPLSAADD317 pKa = 4.44DD318 pKa = 4.46PGLAAGIDD326 pKa = 4.15FSEE329 pKa = 4.9EE330 pKa = 3.83PVDD333 pKa = 3.63YY334 pKa = 10.84CAARR338 pKa = 11.84PVSTASGTFVCASTQLFASRR358 pKa = 11.84ILVLGSLLDD367 pKa = 3.72GEE369 pKa = 5.07AWHH372 pKa = 6.82TDD374 pKa = 2.5ISEE377 pKa = 4.12DD378 pKa = 3.36AFEE381 pKa = 4.95FGLFNEE387 pKa = 4.76QYY389 pKa = 10.88GRR391 pKa = 11.84LHH393 pKa = 6.19TPVDD397 pKa = 3.74GTDD400 pKa = 3.32APLVIMPGLDD410 pKa = 2.8AGVRR414 pKa = 11.84AVSAVDD420 pKa = 5.48GSTLWEE426 pKa = 4.49AGEE429 pKa = 4.87DD430 pKa = 4.12LDD432 pKa = 5.44PLDD435 pKa = 4.7PVEE438 pKa = 4.24GSYY441 pKa = 11.58GFGGNGAFPQLGLIPVLLNDD461 pKa = 3.64AVHH464 pKa = 7.02FFTLDD469 pKa = 3.44GKK471 pKa = 9.95EE472 pKa = 3.98HH473 pKa = 6.86AEE475 pKa = 3.76AHH477 pKa = 6.22AVGQYY482 pKa = 10.7AEE484 pKa = 4.54PSATGRR490 pKa = 11.84WFAAAAGEE498 pKa = 4.52STTLWTVTGGG508 pKa = 3.29
Molecular weight: 52.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N6PGI4|A0A2N6PGI4_9MICO NUDIX hydrolase OS=Brevibacterium luteolum OX=199591 GN=CJ198_09325 PE=4 SV=1
MM1 pKa = 7.56SKK3 pKa = 8.12HH4 pKa = 5.4TFQPNNRR11 pKa = 11.84KK12 pKa = 9.39RR13 pKa = 11.84SKK15 pKa = 10.21KK16 pKa = 9.28HH17 pKa = 4.28GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.16GRR40 pKa = 11.84KK41 pKa = 8.86ALSAA45 pKa = 3.99
MM1 pKa = 7.56SKK3 pKa = 8.12HH4 pKa = 5.4TFQPNNRR11 pKa = 11.84KK12 pKa = 9.39RR13 pKa = 11.84SKK15 pKa = 10.21KK16 pKa = 9.28HH17 pKa = 4.28GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.16GRR40 pKa = 11.84KK41 pKa = 8.86ALSAA45 pKa = 3.99
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
903331 |
32 |
2678 |
332.7 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.069 ± 0.073 | 0.72 ± 0.011 |
6.262 ± 0.046 | 6.181 ± 0.045 |
3.104 ± 0.027 | 8.704 ± 0.042 |
2.192 ± 0.022 | 4.938 ± 0.034 |
2.488 ± 0.038 | 9.639 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.07 ± 0.019 | 2.092 ± 0.026 |
5.339 ± 0.032 | 3.173 ± 0.028 |
6.756 ± 0.046 | 5.74 ± 0.033 |
6.333 ± 0.037 | 7.87 ± 0.037 |
1.319 ± 0.02 | 2.011 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
