
Yersinia phage fPS-54-ocr
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Helsettvirus; Yersinia virus fPS54ocr
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
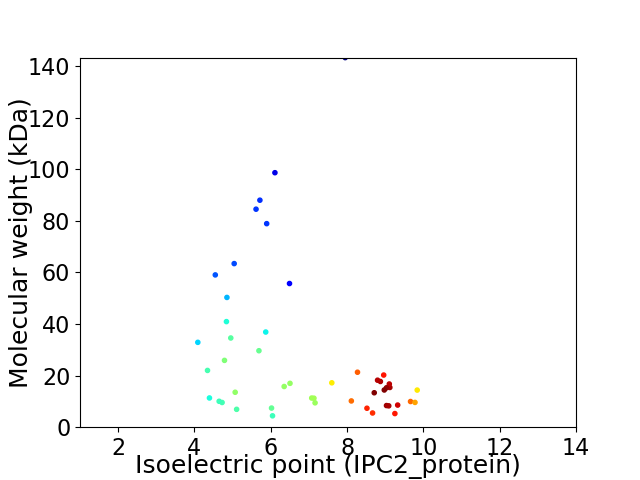
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H1UJF1|A0A2H1UJF1_9CAUD Phage endopeptidase RZ1 OS=Yersinia phage fPS-54-ocr OX=2052753 GN=g042 PE=4 SV=1
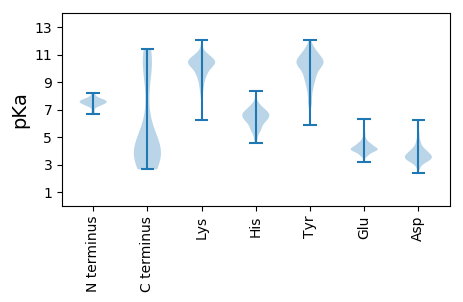
MM1 pKa = 7.47SNEE4 pKa = 4.05SNADD8 pKa = 3.08VYY10 pKa = 11.65AEE12 pKa = 4.07MGANPTAVTGSMSDD26 pKa = 3.25HH27 pKa = 6.52DD28 pKa = 4.13TSMLEE33 pKa = 3.92LDD35 pKa = 3.41VDD37 pKa = 3.97TRR39 pKa = 11.84DD40 pKa = 3.24GDD42 pKa = 4.18DD43 pKa = 5.06RR44 pKa = 11.84ITLVDD49 pKa = 4.48NDD51 pKa = 4.38DD52 pKa = 4.49PYY54 pKa = 11.39NTVDD58 pKa = 3.75PFADD62 pKa = 3.88PEE64 pKa = 5.51ADD66 pKa = 3.49DD67 pKa = 3.8GTRR70 pKa = 11.84NSLRR74 pKa = 11.84ISSEE78 pKa = 3.61GNEE81 pKa = 4.94DD82 pKa = 3.17IDD84 pKa = 4.47SGEE87 pKa = 4.24GEE89 pKa = 4.06PDD91 pKa = 3.05EE92 pKa = 4.65SQSEE96 pKa = 4.17EE97 pKa = 3.93ATDD100 pKa = 4.47FEE102 pKa = 6.3AIGDD106 pKa = 4.04APSEE110 pKa = 4.06LTEE113 pKa = 4.56ASAQLEE119 pKa = 4.08EE120 pKa = 4.9HH121 pKa = 6.18EE122 pKa = 5.51AGFQEE127 pKa = 4.63MVDD130 pKa = 3.49AAAEE134 pKa = 4.11RR135 pKa = 11.84GLSDD139 pKa = 4.66DD140 pKa = 4.94AITRR144 pKa = 11.84IQSEE148 pKa = 4.46YY149 pKa = 10.69QGDD152 pKa = 4.14GLTKK156 pKa = 10.32EE157 pKa = 4.52SYY159 pKa = 10.72EE160 pKa = 3.99EE161 pKa = 3.79LAAAGYY167 pKa = 9.99SKK169 pKa = 11.12SFVDD173 pKa = 3.98SYY175 pKa = 11.69IRR177 pKa = 11.84GQEE180 pKa = 3.98ALVEE184 pKa = 4.54SYY186 pKa = 10.87VKK188 pKa = 10.61SVFAYY193 pKa = 10.29AGGEE197 pKa = 3.99QQFNALYY204 pKa = 9.04THH206 pKa = 7.16LEE208 pKa = 3.96ATNAEE213 pKa = 4.25AAQSLVSALEE223 pKa = 3.9SRR225 pKa = 11.84DD226 pKa = 3.44LSTVKK231 pKa = 10.73AIVNLAGASRR241 pKa = 11.84AKK243 pKa = 10.23TFGKK247 pKa = 9.29PAARR251 pKa = 11.84SVTKK255 pKa = 9.17QARR258 pKa = 11.84QSAPVSNKK266 pKa = 8.71VVGYY270 pKa = 8.48TNSADD275 pKa = 3.39MMKK278 pKa = 10.98DD279 pKa = 3.14MNDD282 pKa = 3.06PRR284 pKa = 11.84YY285 pKa = 9.39STDD288 pKa = 2.59AKK290 pKa = 10.31FRR292 pKa = 11.84KK293 pKa = 9.26EE294 pKa = 4.06VEE296 pKa = 4.2LKK298 pKa = 10.05TMYY301 pKa = 9.18STYY304 pKa = 11.44
MM1 pKa = 7.47SNEE4 pKa = 4.05SNADD8 pKa = 3.08VYY10 pKa = 11.65AEE12 pKa = 4.07MGANPTAVTGSMSDD26 pKa = 3.25HH27 pKa = 6.52DD28 pKa = 4.13TSMLEE33 pKa = 3.92LDD35 pKa = 3.41VDD37 pKa = 3.97TRR39 pKa = 11.84DD40 pKa = 3.24GDD42 pKa = 4.18DD43 pKa = 5.06RR44 pKa = 11.84ITLVDD49 pKa = 4.48NDD51 pKa = 4.38DD52 pKa = 4.49PYY54 pKa = 11.39NTVDD58 pKa = 3.75PFADD62 pKa = 3.88PEE64 pKa = 5.51ADD66 pKa = 3.49DD67 pKa = 3.8GTRR70 pKa = 11.84NSLRR74 pKa = 11.84ISSEE78 pKa = 3.61GNEE81 pKa = 4.94DD82 pKa = 3.17IDD84 pKa = 4.47SGEE87 pKa = 4.24GEE89 pKa = 4.06PDD91 pKa = 3.05EE92 pKa = 4.65SQSEE96 pKa = 4.17EE97 pKa = 3.93ATDD100 pKa = 4.47FEE102 pKa = 6.3AIGDD106 pKa = 4.04APSEE110 pKa = 4.06LTEE113 pKa = 4.56ASAQLEE119 pKa = 4.08EE120 pKa = 4.9HH121 pKa = 6.18EE122 pKa = 5.51AGFQEE127 pKa = 4.63MVDD130 pKa = 3.49AAAEE134 pKa = 4.11RR135 pKa = 11.84GLSDD139 pKa = 4.66DD140 pKa = 4.94AITRR144 pKa = 11.84IQSEE148 pKa = 4.46YY149 pKa = 10.69QGDD152 pKa = 4.14GLTKK156 pKa = 10.32EE157 pKa = 4.52SYY159 pKa = 10.72EE160 pKa = 3.99EE161 pKa = 3.79LAAAGYY167 pKa = 9.99SKK169 pKa = 11.12SFVDD173 pKa = 3.98SYY175 pKa = 11.69IRR177 pKa = 11.84GQEE180 pKa = 3.98ALVEE184 pKa = 4.54SYY186 pKa = 10.87VKK188 pKa = 10.61SVFAYY193 pKa = 10.29AGGEE197 pKa = 3.99QQFNALYY204 pKa = 9.04THH206 pKa = 7.16LEE208 pKa = 3.96ATNAEE213 pKa = 4.25AAQSLVSALEE223 pKa = 3.9SRR225 pKa = 11.84DD226 pKa = 3.44LSTVKK231 pKa = 10.73AIVNLAGASRR241 pKa = 11.84AKK243 pKa = 10.23TFGKK247 pKa = 9.29PAARR251 pKa = 11.84SVTKK255 pKa = 9.17QARR258 pKa = 11.84QSAPVSNKK266 pKa = 8.71VVGYY270 pKa = 8.48TNSADD275 pKa = 3.39MMKK278 pKa = 10.98DD279 pKa = 3.14MNDD282 pKa = 3.06PRR284 pKa = 11.84YY285 pKa = 9.39STDD288 pKa = 2.59AKK290 pKa = 10.31FRR292 pKa = 11.84KK293 pKa = 9.26EE294 pKa = 4.06VEE296 pKa = 4.2LKK298 pKa = 10.05TMYY301 pKa = 9.18STYY304 pKa = 11.44
Molecular weight: 32.91 kDa
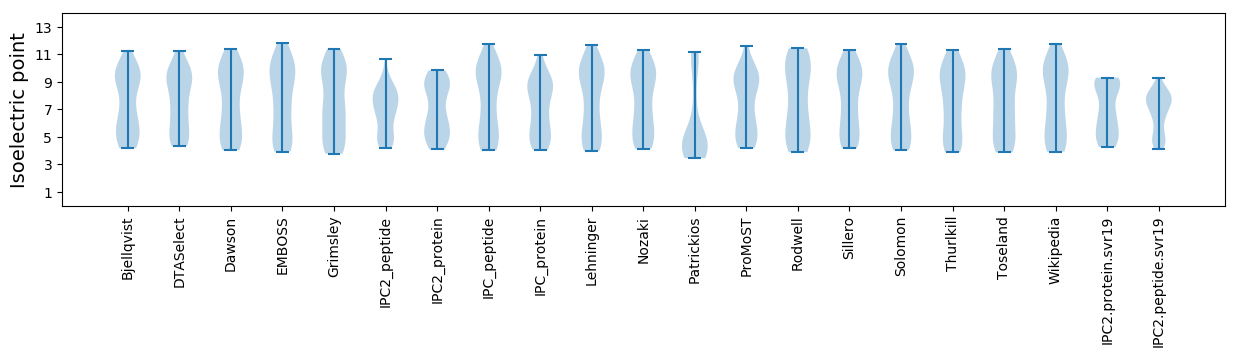
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H1UJF4|A0A2H1UJF4_9CAUD Uncharacterized protein OS=Yersinia phage fPS-54-ocr OX=2052753 GN=g045 PE=4 SV=1
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.46SYY5 pKa = 10.37KK6 pKa = 9.87QFYY9 pKa = 8.47KK10 pKa = 10.46QPRR13 pKa = 11.84NHH15 pKa = 5.49IQVWVSANGPIPSGYY30 pKa = 10.51YY31 pKa = 9.08IDD33 pKa = 5.89HH34 pKa = 7.39IDD36 pKa = 4.66GNPLNDD42 pKa = 3.55NLDD45 pKa = 3.94NLRR48 pKa = 11.84LATPSEE54 pKa = 4.02NSRR57 pKa = 11.84NMKK60 pKa = 9.28TPSSNITGLKK70 pKa = 7.7WLSWCPLRR78 pKa = 11.84LTWRR82 pKa = 11.84GTLKK86 pKa = 10.8VNYY89 pKa = 8.48KK90 pKa = 7.42QHH92 pKa = 6.43SYY94 pKa = 10.55RR95 pKa = 11.84SKK97 pKa = 11.16DD98 pKa = 3.43LFDD101 pKa = 3.6VVSWLFRR108 pKa = 11.84TRR110 pKa = 11.84RR111 pKa = 11.84EE112 pKa = 3.5IHH114 pKa = 5.83GQFARR119 pKa = 11.84NRR121 pKa = 3.65
MM1 pKa = 7.62RR2 pKa = 11.84KK3 pKa = 9.46SYY5 pKa = 10.37KK6 pKa = 9.87QFYY9 pKa = 8.47KK10 pKa = 10.46QPRR13 pKa = 11.84NHH15 pKa = 5.49IQVWVSANGPIPSGYY30 pKa = 10.51YY31 pKa = 9.08IDD33 pKa = 5.89HH34 pKa = 7.39IDD36 pKa = 4.66GNPLNDD42 pKa = 3.55NLDD45 pKa = 3.94NLRR48 pKa = 11.84LATPSEE54 pKa = 4.02NSRR57 pKa = 11.84NMKK60 pKa = 9.28TPSSNITGLKK70 pKa = 7.7WLSWCPLRR78 pKa = 11.84LTWRR82 pKa = 11.84GTLKK86 pKa = 10.8VNYY89 pKa = 8.48KK90 pKa = 7.42QHH92 pKa = 6.43SYY94 pKa = 10.55RR95 pKa = 11.84SKK97 pKa = 11.16DD98 pKa = 3.43LFDD101 pKa = 3.6VVSWLFRR108 pKa = 11.84TRR110 pKa = 11.84RR111 pKa = 11.84EE112 pKa = 3.5IHH114 pKa = 5.83GQFARR119 pKa = 11.84NRR121 pKa = 3.65
Molecular weight: 14.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11936 |
41 |
1317 |
248.7 |
27.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.638 ± 0.483 | 0.938 ± 0.143 |
6.334 ± 0.274 | 6.744 ± 0.282 |
3.452 ± 0.171 | 7.666 ± 0.252 |
1.785 ± 0.176 | 5.312 ± 0.225 |
6.82 ± 0.365 | 8.068 ± 0.275 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.983 ± 0.176 | 4.708 ± 0.266 |
3.544 ± 0.225 | 3.795 ± 0.25 |
5.546 ± 0.198 | 6.409 ± 0.324 |
5.747 ± 0.196 | 6.635 ± 0.252 |
1.424 ± 0.165 | 3.452 ± 0.203 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
