
Sinapis alba cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
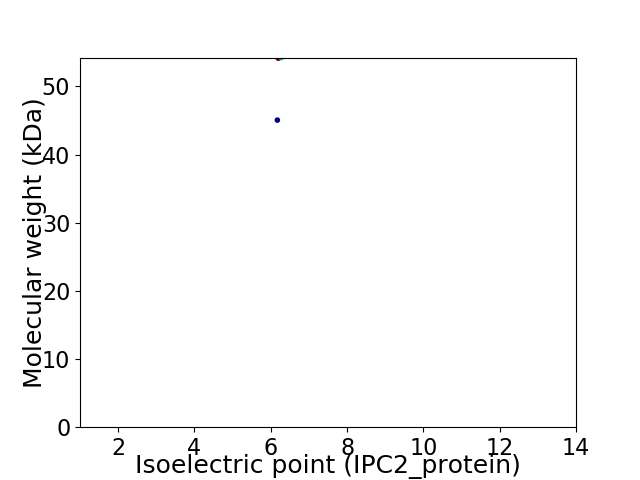
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U3AHS1|A0A0U3AHS1_9VIRU Capsid protein OS=Sinapis alba cryptic virus 1 OX=1768874 PE=4 SV=1
MM1 pKa = 6.52TTGDD5 pKa = 3.87RR6 pKa = 11.84VTPTDD11 pKa = 4.7GITTVDD17 pKa = 3.63APTATLEE24 pKa = 4.21AKK26 pKa = 10.33LDD28 pKa = 3.96SSSTAYY34 pKa = 10.46LKK36 pKa = 10.85SATDD40 pKa = 4.05FLTQSSRR47 pKa = 11.84IGTLRR52 pKa = 11.84TSYY55 pKa = 10.77KK56 pKa = 9.84DD57 pKa = 3.04RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 10.25VYY63 pKa = 10.56LKK65 pKa = 9.52SQAMFDD71 pKa = 4.05SLVEE75 pKa = 4.22LYY77 pKa = 10.89SLLFTTQWQVFRR89 pKa = 11.84RR90 pKa = 11.84FTDD93 pKa = 3.12IFVPPANVTATRR105 pKa = 11.84YY106 pKa = 9.92LAMNYY111 pKa = 9.6ISMWIQDD118 pKa = 3.9TYY120 pKa = 11.75AMNRR124 pKa = 11.84AALMKK129 pKa = 10.52LSVVALNDD137 pKa = 3.63RR138 pKa = 11.84FPVEE142 pKa = 4.08IPIVGYY148 pKa = 10.64QYY150 pKa = 11.25DD151 pKa = 3.96AYY153 pKa = 10.65LAHH156 pKa = 7.22LSAALRR162 pKa = 11.84PTLTKK167 pKa = 10.98HH168 pKa = 5.5MMEE171 pKa = 4.18DD172 pKa = 3.99TLWIPCFTRR181 pKa = 11.84SPNFQVEE188 pKa = 4.74SPFGITGYY196 pKa = 10.38VINDD200 pKa = 3.63VIFTGAMNIMKK211 pKa = 9.62EE212 pKa = 3.89RR213 pKa = 11.84KK214 pKa = 7.94LWRR217 pKa = 11.84IIDD220 pKa = 3.92LPTNVLGRR228 pKa = 11.84PSWLFDD234 pKa = 2.91WHH236 pKa = 7.56DD237 pKa = 4.23DD238 pKa = 4.37DD239 pKa = 6.33RR240 pKa = 11.84VCAWFPMEE248 pKa = 5.61GNFTMDD254 pKa = 4.56DD255 pKa = 3.06VTMAYY260 pKa = 9.43ILGVACTPKK269 pKa = 10.49LAPCDD274 pKa = 3.72VDD276 pKa = 3.14DD277 pKa = 4.05WQYY280 pKa = 11.13FPNDD284 pKa = 3.63VVPANLDD291 pKa = 3.17PYY293 pKa = 11.64AFLRR297 pKa = 11.84LTNRR301 pKa = 11.84RR302 pKa = 11.84FHH304 pKa = 6.56GGYY307 pKa = 10.32EE308 pKa = 3.86IRR310 pKa = 11.84TLDD313 pKa = 3.74AANMPGQTGTTTDD326 pKa = 2.66SRR328 pKa = 11.84KK329 pKa = 9.66RR330 pKa = 11.84PRR332 pKa = 11.84PGEE335 pKa = 4.22STSAPALPQTSEE347 pKa = 4.43PQTQMQVDD355 pKa = 4.76TPPAPKK361 pKa = 9.79PRR363 pKa = 11.84FRR365 pKa = 11.84LVDD368 pKa = 3.15WTYY371 pKa = 11.16HH372 pKa = 5.06YY373 pKa = 11.24LVVLNKK379 pKa = 9.85EE380 pKa = 3.78DD381 pKa = 3.49HH382 pKa = 6.28TRR384 pKa = 11.84FAALKK389 pKa = 9.08SICWNN394 pKa = 3.46
MM1 pKa = 6.52TTGDD5 pKa = 3.87RR6 pKa = 11.84VTPTDD11 pKa = 4.7GITTVDD17 pKa = 3.63APTATLEE24 pKa = 4.21AKK26 pKa = 10.33LDD28 pKa = 3.96SSSTAYY34 pKa = 10.46LKK36 pKa = 10.85SATDD40 pKa = 4.05FLTQSSRR47 pKa = 11.84IGTLRR52 pKa = 11.84TSYY55 pKa = 10.77KK56 pKa = 9.84DD57 pKa = 3.04RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 10.25VYY63 pKa = 10.56LKK65 pKa = 9.52SQAMFDD71 pKa = 4.05SLVEE75 pKa = 4.22LYY77 pKa = 10.89SLLFTTQWQVFRR89 pKa = 11.84RR90 pKa = 11.84FTDD93 pKa = 3.12IFVPPANVTATRR105 pKa = 11.84YY106 pKa = 9.92LAMNYY111 pKa = 9.6ISMWIQDD118 pKa = 3.9TYY120 pKa = 11.75AMNRR124 pKa = 11.84AALMKK129 pKa = 10.52LSVVALNDD137 pKa = 3.63RR138 pKa = 11.84FPVEE142 pKa = 4.08IPIVGYY148 pKa = 10.64QYY150 pKa = 11.25DD151 pKa = 3.96AYY153 pKa = 10.65LAHH156 pKa = 7.22LSAALRR162 pKa = 11.84PTLTKK167 pKa = 10.98HH168 pKa = 5.5MMEE171 pKa = 4.18DD172 pKa = 3.99TLWIPCFTRR181 pKa = 11.84SPNFQVEE188 pKa = 4.74SPFGITGYY196 pKa = 10.38VINDD200 pKa = 3.63VIFTGAMNIMKK211 pKa = 9.62EE212 pKa = 3.89RR213 pKa = 11.84KK214 pKa = 7.94LWRR217 pKa = 11.84IIDD220 pKa = 3.92LPTNVLGRR228 pKa = 11.84PSWLFDD234 pKa = 2.91WHH236 pKa = 7.56DD237 pKa = 4.23DD238 pKa = 4.37DD239 pKa = 6.33RR240 pKa = 11.84VCAWFPMEE248 pKa = 5.61GNFTMDD254 pKa = 4.56DD255 pKa = 3.06VTMAYY260 pKa = 9.43ILGVACTPKK269 pKa = 10.49LAPCDD274 pKa = 3.72VDD276 pKa = 3.14DD277 pKa = 4.05WQYY280 pKa = 11.13FPNDD284 pKa = 3.63VVPANLDD291 pKa = 3.17PYY293 pKa = 11.64AFLRR297 pKa = 11.84LTNRR301 pKa = 11.84RR302 pKa = 11.84FHH304 pKa = 6.56GGYY307 pKa = 10.32EE308 pKa = 3.86IRR310 pKa = 11.84TLDD313 pKa = 3.74AANMPGQTGTTTDD326 pKa = 2.66SRR328 pKa = 11.84KK329 pKa = 9.66RR330 pKa = 11.84PRR332 pKa = 11.84PGEE335 pKa = 4.22STSAPALPQTSEE347 pKa = 4.43PQTQMQVDD355 pKa = 4.76TPPAPKK361 pKa = 9.79PRR363 pKa = 11.84FRR365 pKa = 11.84LVDD368 pKa = 3.15WTYY371 pKa = 11.16HH372 pKa = 5.06YY373 pKa = 11.24LVVLNKK379 pKa = 9.85EE380 pKa = 3.78DD381 pKa = 3.49HH382 pKa = 6.28TRR384 pKa = 11.84FAALKK389 pKa = 9.08SICWNN394 pKa = 3.46
Molecular weight: 45.06 kDa
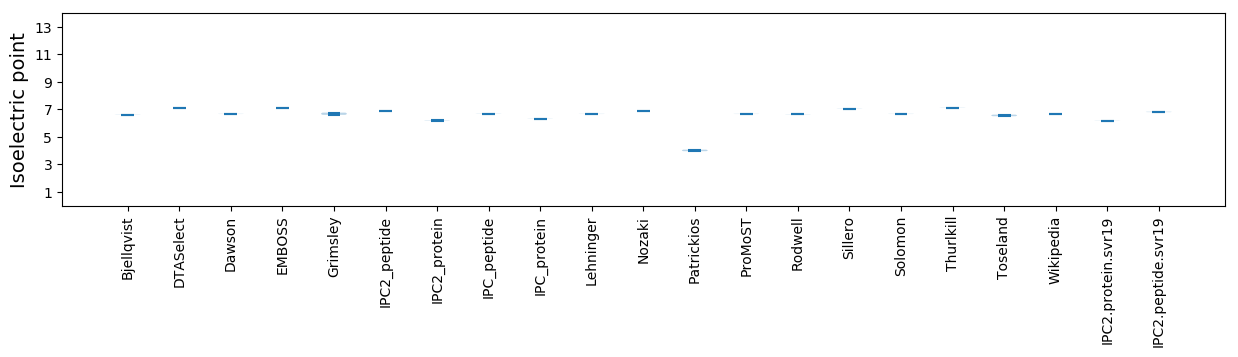
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U3AHS1|A0A0U3AHS1_9VIRU Capsid protein OS=Sinapis alba cryptic virus 1 OX=1768874 PE=4 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84PSITGYY8 pKa = 10.77DD9 pKa = 3.66YY10 pKa = 11.72TNFTQDD16 pKa = 3.65LLKK19 pKa = 10.39SDD21 pKa = 4.06RR22 pKa = 11.84KK23 pKa = 9.53HH24 pKa = 5.14PHH26 pKa = 3.57VVRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.7TATTYY36 pKa = 11.01RR37 pKa = 11.84DD38 pKa = 3.3DD39 pKa = 3.86FAFKK43 pKa = 10.48EE44 pKa = 4.54VISLDD49 pKa = 3.07RR50 pKa = 11.84LAYY53 pKa = 8.99IQRR56 pKa = 11.84LEE58 pKa = 3.95GWSRR62 pKa = 11.84SYY64 pKa = 11.09YY65 pKa = 10.56LPEE68 pKa = 3.89KK69 pKa = 10.34HH70 pKa = 6.85LEE72 pKa = 4.03ALLQYY77 pKa = 7.38ATPNVPCTALNLNVYY92 pKa = 9.02RR93 pKa = 11.84QAIQVVEE100 pKa = 4.0NGLRR104 pKa = 11.84SLQPVRR110 pKa = 11.84AFDD113 pKa = 4.02VLTEE117 pKa = 4.46LNQISYY123 pKa = 9.09KK124 pKa = 9.82QSSAAGYY131 pKa = 10.32DD132 pKa = 3.61YY133 pKa = 10.87IGAKK137 pKa = 10.36GPIDD141 pKa = 3.9GEE143 pKa = 4.05NHH145 pKa = 5.27KK146 pKa = 10.57RR147 pKa = 11.84AISRR151 pKa = 11.84AKK153 pKa = 9.39AVLWSVVKK161 pKa = 10.64EE162 pKa = 4.03DD163 pKa = 5.04GEE165 pKa = 4.89GIDD168 pKa = 3.87HH169 pKa = 7.55AIEE172 pKa = 4.02TSVPDD177 pKa = 3.16VGYY180 pKa = 8.78TRR182 pKa = 11.84TQLADD187 pKa = 3.48LTEE190 pKa = 4.21KK191 pKa = 10.41TKK193 pKa = 10.33VRR195 pKa = 11.84QVWGRR200 pKa = 11.84AFHH203 pKa = 6.55YY204 pKa = 10.25ILLEE208 pKa = 4.02GLVAQPFIQSIMEE221 pKa = 4.36GPSFIHH227 pKa = 6.66TGRR230 pKa = 11.84DD231 pKa = 3.27PTLSVPQSLAKK242 pKa = 10.57VSSQCKK248 pKa = 9.75YY249 pKa = 10.34IYY251 pKa = 10.89SLDD254 pKa = 3.53WKK256 pKa = 11.19SFDD259 pKa = 3.12ATVNRR264 pKa = 11.84FEE266 pKa = 5.28INTSFDD272 pKa = 3.5IIKK275 pKa = 10.57SKK277 pKa = 11.16VIFPNYY283 pKa = 7.0EE284 pKa = 3.55TEE286 pKa = 3.98QAFEE290 pKa = 3.82ITRR293 pKa = 11.84QLFLHH298 pKa = 6.4KK299 pKa = 10.14KK300 pKa = 8.12VAAPDD305 pKa = 4.08GYY307 pKa = 10.13IYY309 pKa = 9.6EE310 pKa = 4.17AHH312 pKa = 6.84KK313 pKa = 10.56GIPSGSYY320 pKa = 6.51YY321 pKa = 10.55TSMVGSIVNRR331 pKa = 11.84LRR333 pKa = 11.84IEE335 pKa = 4.36YY336 pKa = 9.01IWRR339 pKa = 11.84IATGHH344 pKa = 6.11GPIHH348 pKa = 7.12CEE350 pKa = 3.71TLGDD354 pKa = 4.95DD355 pKa = 4.45SLCGDD360 pKa = 5.25DD361 pKa = 4.23IFVPATQLADD371 pKa = 2.98IANRR375 pKa = 11.84IGWYY379 pKa = 10.2FNADD383 pKa = 2.91KK384 pKa = 10.71TEE386 pKa = 3.87YY387 pKa = 9.31STIPEE392 pKa = 4.2GVTFLGRR399 pKa = 11.84TSTGNLNSRR408 pKa = 11.84DD409 pKa = 3.43LTKK412 pKa = 10.61CLRR415 pKa = 11.84LLVYY419 pKa = 9.82PEE421 pKa = 4.27YY422 pKa = 9.97PVTSGRR428 pKa = 11.84ISAYY432 pKa = 9.12RR433 pKa = 11.84ARR435 pKa = 11.84SIADD439 pKa = 3.41DD440 pKa = 3.77SGGLSDD446 pKa = 6.05LLNQVAIRR454 pKa = 11.84LEE456 pKa = 3.93RR457 pKa = 11.84SYY459 pKa = 11.53GIASEE464 pKa = 4.48EE465 pKa = 4.05EE466 pKa = 3.45IPAYY470 pKa = 9.91FKK472 pKa = 10.74RR473 pKa = 11.84YY474 pKa = 8.94VPFMM478 pKa = 4.76
MM1 pKa = 7.6RR2 pKa = 11.84PSITGYY8 pKa = 10.77DD9 pKa = 3.66YY10 pKa = 11.72TNFTQDD16 pKa = 3.65LLKK19 pKa = 10.39SDD21 pKa = 4.06RR22 pKa = 11.84KK23 pKa = 9.53HH24 pKa = 5.14PHH26 pKa = 3.57VVRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.7TATTYY36 pKa = 11.01RR37 pKa = 11.84DD38 pKa = 3.3DD39 pKa = 3.86FAFKK43 pKa = 10.48EE44 pKa = 4.54VISLDD49 pKa = 3.07RR50 pKa = 11.84LAYY53 pKa = 8.99IQRR56 pKa = 11.84LEE58 pKa = 3.95GWSRR62 pKa = 11.84SYY64 pKa = 11.09YY65 pKa = 10.56LPEE68 pKa = 3.89KK69 pKa = 10.34HH70 pKa = 6.85LEE72 pKa = 4.03ALLQYY77 pKa = 7.38ATPNVPCTALNLNVYY92 pKa = 9.02RR93 pKa = 11.84QAIQVVEE100 pKa = 4.0NGLRR104 pKa = 11.84SLQPVRR110 pKa = 11.84AFDD113 pKa = 4.02VLTEE117 pKa = 4.46LNQISYY123 pKa = 9.09KK124 pKa = 9.82QSSAAGYY131 pKa = 10.32DD132 pKa = 3.61YY133 pKa = 10.87IGAKK137 pKa = 10.36GPIDD141 pKa = 3.9GEE143 pKa = 4.05NHH145 pKa = 5.27KK146 pKa = 10.57RR147 pKa = 11.84AISRR151 pKa = 11.84AKK153 pKa = 9.39AVLWSVVKK161 pKa = 10.64EE162 pKa = 4.03DD163 pKa = 5.04GEE165 pKa = 4.89GIDD168 pKa = 3.87HH169 pKa = 7.55AIEE172 pKa = 4.02TSVPDD177 pKa = 3.16VGYY180 pKa = 8.78TRR182 pKa = 11.84TQLADD187 pKa = 3.48LTEE190 pKa = 4.21KK191 pKa = 10.41TKK193 pKa = 10.33VRR195 pKa = 11.84QVWGRR200 pKa = 11.84AFHH203 pKa = 6.55YY204 pKa = 10.25ILLEE208 pKa = 4.02GLVAQPFIQSIMEE221 pKa = 4.36GPSFIHH227 pKa = 6.66TGRR230 pKa = 11.84DD231 pKa = 3.27PTLSVPQSLAKK242 pKa = 10.57VSSQCKK248 pKa = 9.75YY249 pKa = 10.34IYY251 pKa = 10.89SLDD254 pKa = 3.53WKK256 pKa = 11.19SFDD259 pKa = 3.12ATVNRR264 pKa = 11.84FEE266 pKa = 5.28INTSFDD272 pKa = 3.5IIKK275 pKa = 10.57SKK277 pKa = 11.16VIFPNYY283 pKa = 7.0EE284 pKa = 3.55TEE286 pKa = 3.98QAFEE290 pKa = 3.82ITRR293 pKa = 11.84QLFLHH298 pKa = 6.4KK299 pKa = 10.14KK300 pKa = 8.12VAAPDD305 pKa = 4.08GYY307 pKa = 10.13IYY309 pKa = 9.6EE310 pKa = 4.17AHH312 pKa = 6.84KK313 pKa = 10.56GIPSGSYY320 pKa = 6.51YY321 pKa = 10.55TSMVGSIVNRR331 pKa = 11.84LRR333 pKa = 11.84IEE335 pKa = 4.36YY336 pKa = 9.01IWRR339 pKa = 11.84IATGHH344 pKa = 6.11GPIHH348 pKa = 7.12CEE350 pKa = 3.71TLGDD354 pKa = 4.95DD355 pKa = 4.45SLCGDD360 pKa = 5.25DD361 pKa = 4.23IFVPATQLADD371 pKa = 2.98IANRR375 pKa = 11.84IGWYY379 pKa = 10.2FNADD383 pKa = 2.91KK384 pKa = 10.71TEE386 pKa = 3.87YY387 pKa = 9.31STIPEE392 pKa = 4.2GVTFLGRR399 pKa = 11.84TSTGNLNSRR408 pKa = 11.84DD409 pKa = 3.43LTKK412 pKa = 10.61CLRR415 pKa = 11.84LLVYY419 pKa = 9.82PEE421 pKa = 4.27YY422 pKa = 9.97PVTSGRR428 pKa = 11.84ISAYY432 pKa = 9.12RR433 pKa = 11.84ARR435 pKa = 11.84SIADD439 pKa = 3.41DD440 pKa = 3.77SGGLSDD446 pKa = 6.05LLNQVAIRR454 pKa = 11.84LEE456 pKa = 3.93RR457 pKa = 11.84SYY459 pKa = 11.53GIASEE464 pKa = 4.48EE465 pKa = 4.05EE466 pKa = 3.45IPAYY470 pKa = 9.91FKK472 pKa = 10.74RR473 pKa = 11.84YY474 pKa = 8.94VPFMM478 pKa = 4.76
Molecular weight: 54.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872 |
394 |
478 |
436.0 |
49.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.569 ± 0.186 | 1.147 ± 0.076 |
6.651 ± 0.598 | 4.472 ± 1.044 |
4.243 ± 0.36 | 5.161 ± 0.841 |
1.95 ± 0.265 | 6.078 ± 1.096 |
4.014 ± 0.444 | 8.486 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.179 ± 1.012 | 3.555 ± 0.157 |
5.734 ± 0.853 | 3.555 ± 0.159 |
6.766 ± 0.054 | 6.651 ± 0.821 |
8.257 ± 1.335 | 6.307 ± 0.024 |
1.835 ± 0.437 | 5.39 ± 0.51 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
