
Wuhan insect virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
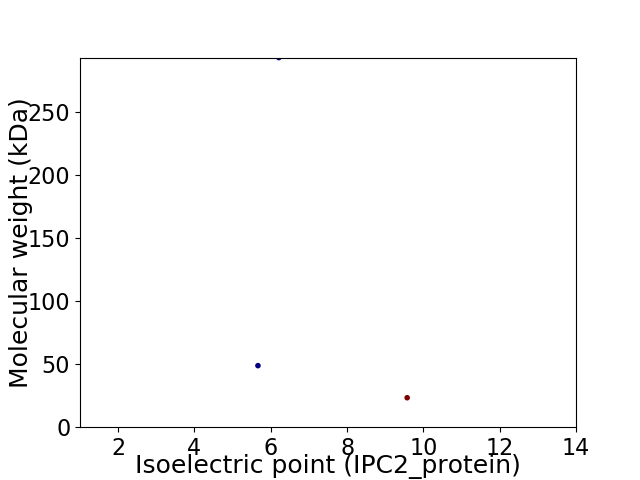
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK77|A0A1L3KK77_9VIRU Uncharacterized protein OS=Wuhan insect virus 8 OX=1923739 PE=4 SV=1
MM1 pKa = 7.31SSSYY5 pKa = 10.71FITLFSLILLSLYY18 pKa = 10.25LCKK21 pKa = 10.6NFQNVSSSQLLSTFYY36 pKa = 11.12NSSFWNKK43 pKa = 9.85LGRR46 pKa = 11.84SASISNHH53 pKa = 6.75PINEE57 pKa = 4.03YY58 pKa = 10.74LSAKK62 pKa = 9.98ARR64 pKa = 11.84LDD66 pKa = 3.51LLGTYY71 pKa = 9.85SSSVFSFDD79 pKa = 3.61RR80 pKa = 11.84EE81 pKa = 4.36CISVVKK87 pKa = 9.72TNALLEE93 pKa = 4.29VDD95 pKa = 3.99CQLPSRR101 pKa = 11.84CGTLHH106 pKa = 6.95HH107 pKa = 6.92KK108 pKa = 10.33LLSKK112 pKa = 9.44TFYY115 pKa = 8.35GQEE118 pKa = 3.65VRR120 pKa = 11.84VCYY123 pKa = 10.29SFHH126 pKa = 6.68HH127 pKa = 6.06SVYY130 pKa = 10.99ADD132 pKa = 4.66AINLYY137 pKa = 8.45TIKK140 pKa = 10.46FPRR143 pKa = 11.84DD144 pKa = 3.09LHH146 pKa = 7.88YY147 pKa = 11.54GMLTQHH153 pKa = 7.07DD154 pKa = 4.75SVDD157 pKa = 3.29VHH159 pKa = 7.29DD160 pKa = 4.82IVPVDD165 pKa = 3.98FFEE168 pKa = 6.13HH169 pKa = 6.21IVIANATDD177 pKa = 5.17GYY179 pKa = 11.13HH180 pKa = 7.31LFPQICLQQYY190 pKa = 8.3TSYY193 pKa = 11.54SDD195 pKa = 4.7DD196 pKa = 4.1LPPSVVFKK204 pKa = 11.01EE205 pKa = 4.34SDD207 pKa = 3.27DD208 pKa = 4.0TLCVSHH214 pKa = 6.96TSLANVDD221 pKa = 3.76CRR223 pKa = 11.84PLFFPSVAAIVTTFDD238 pKa = 3.15FPIAFKK244 pKa = 10.83YY245 pKa = 10.24NPYY248 pKa = 10.89NYY250 pKa = 10.22LLHH253 pKa = 6.93LKK255 pKa = 9.8FRR257 pKa = 11.84NQDD260 pKa = 3.23PDD262 pKa = 3.64VFGYY266 pKa = 10.75AADD269 pKa = 3.69TTRR272 pKa = 11.84GYY274 pKa = 11.06FPISFINNLNYY285 pKa = 10.01VSRR288 pKa = 11.84SPLSSHH294 pKa = 6.71FLDD297 pKa = 4.18RR298 pKa = 11.84NGSSTVFSRR307 pKa = 11.84NYY309 pKa = 9.34IYY311 pKa = 10.57FDD313 pKa = 3.51SRR315 pKa = 11.84VHH317 pKa = 5.95SFSSKK322 pKa = 9.77DD323 pKa = 3.12YY324 pKa = 9.96CYY326 pKa = 10.43RR327 pKa = 11.84IEE329 pKa = 4.35SLYY332 pKa = 10.92KK333 pKa = 10.52NPISVLASTLISVVSPIFYY352 pKa = 10.49EE353 pKa = 4.06LLEE356 pKa = 4.1LLEE359 pKa = 4.81IEE361 pKa = 4.35VEE363 pKa = 4.13NIIRR367 pKa = 11.84LIVKK371 pKa = 10.17AFLFFLKK378 pKa = 10.6LVVEE382 pKa = 4.46LLSLIITPNVLSALMCAFSIYY403 pKa = 10.01IYY405 pKa = 10.76CFDD408 pKa = 3.86FVITFVSFLIFLAFFLSYY426 pKa = 11.19
MM1 pKa = 7.31SSSYY5 pKa = 10.71FITLFSLILLSLYY18 pKa = 10.25LCKK21 pKa = 10.6NFQNVSSSQLLSTFYY36 pKa = 11.12NSSFWNKK43 pKa = 9.85LGRR46 pKa = 11.84SASISNHH53 pKa = 6.75PINEE57 pKa = 4.03YY58 pKa = 10.74LSAKK62 pKa = 9.98ARR64 pKa = 11.84LDD66 pKa = 3.51LLGTYY71 pKa = 9.85SSSVFSFDD79 pKa = 3.61RR80 pKa = 11.84EE81 pKa = 4.36CISVVKK87 pKa = 9.72TNALLEE93 pKa = 4.29VDD95 pKa = 3.99CQLPSRR101 pKa = 11.84CGTLHH106 pKa = 6.95HH107 pKa = 6.92KK108 pKa = 10.33LLSKK112 pKa = 9.44TFYY115 pKa = 8.35GQEE118 pKa = 3.65VRR120 pKa = 11.84VCYY123 pKa = 10.29SFHH126 pKa = 6.68HH127 pKa = 6.06SVYY130 pKa = 10.99ADD132 pKa = 4.66AINLYY137 pKa = 8.45TIKK140 pKa = 10.46FPRR143 pKa = 11.84DD144 pKa = 3.09LHH146 pKa = 7.88YY147 pKa = 11.54GMLTQHH153 pKa = 7.07DD154 pKa = 4.75SVDD157 pKa = 3.29VHH159 pKa = 7.29DD160 pKa = 4.82IVPVDD165 pKa = 3.98FFEE168 pKa = 6.13HH169 pKa = 6.21IVIANATDD177 pKa = 5.17GYY179 pKa = 11.13HH180 pKa = 7.31LFPQICLQQYY190 pKa = 8.3TSYY193 pKa = 11.54SDD195 pKa = 4.7DD196 pKa = 4.1LPPSVVFKK204 pKa = 11.01EE205 pKa = 4.34SDD207 pKa = 3.27DD208 pKa = 4.0TLCVSHH214 pKa = 6.96TSLANVDD221 pKa = 3.76CRR223 pKa = 11.84PLFFPSVAAIVTTFDD238 pKa = 3.15FPIAFKK244 pKa = 10.83YY245 pKa = 10.24NPYY248 pKa = 10.89NYY250 pKa = 10.22LLHH253 pKa = 6.93LKK255 pKa = 9.8FRR257 pKa = 11.84NQDD260 pKa = 3.23PDD262 pKa = 3.64VFGYY266 pKa = 10.75AADD269 pKa = 3.69TTRR272 pKa = 11.84GYY274 pKa = 11.06FPISFINNLNYY285 pKa = 10.01VSRR288 pKa = 11.84SPLSSHH294 pKa = 6.71FLDD297 pKa = 4.18RR298 pKa = 11.84NGSSTVFSRR307 pKa = 11.84NYY309 pKa = 9.34IYY311 pKa = 10.57FDD313 pKa = 3.51SRR315 pKa = 11.84VHH317 pKa = 5.95SFSSKK322 pKa = 9.77DD323 pKa = 3.12YY324 pKa = 9.96CYY326 pKa = 10.43RR327 pKa = 11.84IEE329 pKa = 4.35SLYY332 pKa = 10.92KK333 pKa = 10.52NPISVLASTLISVVSPIFYY352 pKa = 10.49EE353 pKa = 4.06LLEE356 pKa = 4.1LLEE359 pKa = 4.81IEE361 pKa = 4.35VEE363 pKa = 4.13NIIRR367 pKa = 11.84LIVKK371 pKa = 10.17AFLFFLKK378 pKa = 10.6LVVEE382 pKa = 4.46LLSLIITPNVLSALMCAFSIYY403 pKa = 10.01IYY405 pKa = 10.76CFDD408 pKa = 3.86FVITFVSFLIFLAFFLSYY426 pKa = 11.19
Molecular weight: 48.88 kDa
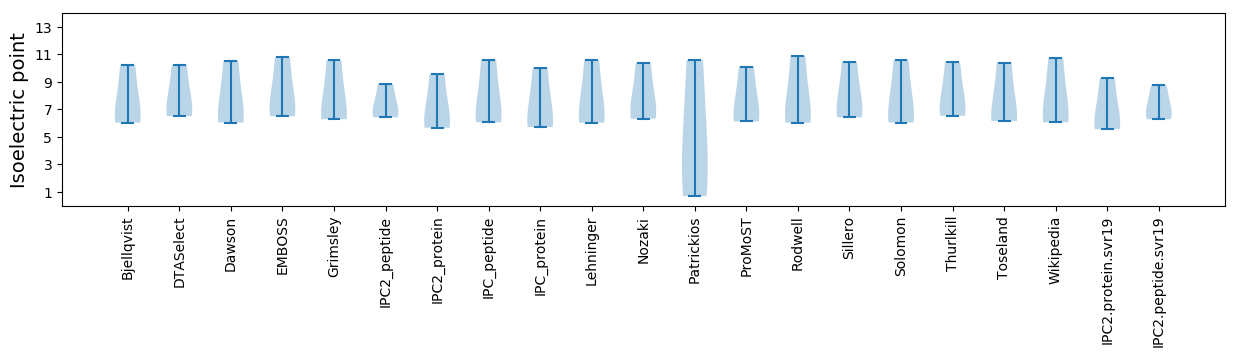
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKD6|A0A1L3KKD6_9VIRU Uncharacterized protein OS=Wuhan insect virus 8 OX=1923739 PE=4 SV=1
MM1 pKa = 7.65ASSEE5 pKa = 4.46SNIGKK10 pKa = 9.99RR11 pKa = 11.84SGNRR15 pKa = 11.84AARR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84NLPRR25 pKa = 11.84NSTVPAVVDD34 pKa = 3.61EE35 pKa = 4.42LTPEE39 pKa = 3.87VSGVLRR45 pKa = 11.84SGSGRR50 pKa = 11.84RR51 pKa = 11.84NVNGIKK57 pKa = 10.26SKK59 pKa = 10.95NKK61 pKa = 10.19SYY63 pKa = 11.35DD64 pKa = 3.44FNQVFSDD71 pKa = 3.28IALTLTSAISNPLVLLTIALSVGVITTHH99 pKa = 6.9NFPQNKK105 pKa = 9.01GFIYY109 pKa = 9.2DD110 pKa = 3.87TFRR113 pKa = 11.84EE114 pKa = 4.23KK115 pKa = 11.15NDD117 pKa = 3.94TISRR121 pKa = 11.84WVVDD125 pKa = 3.43NGQKK129 pKa = 10.12VAGFSIFLPAVVDD142 pKa = 3.7SPRR145 pKa = 11.84NFRR148 pKa = 11.84SVIALTSFLWVMVIPEE164 pKa = 4.3SSVVEE169 pKa = 4.14YY170 pKa = 10.0FLQALAVHH178 pKa = 6.63TYY180 pKa = 10.2FRR182 pKa = 11.84LKK184 pKa = 9.76TDD186 pKa = 3.24NSRR189 pKa = 11.84LTLLAIIGIAWFLGYY204 pKa = 8.88FHH206 pKa = 7.6IPTTKK211 pKa = 10.49
MM1 pKa = 7.65ASSEE5 pKa = 4.46SNIGKK10 pKa = 9.99RR11 pKa = 11.84SGNRR15 pKa = 11.84AARR18 pKa = 11.84RR19 pKa = 11.84SRR21 pKa = 11.84NLPRR25 pKa = 11.84NSTVPAVVDD34 pKa = 3.61EE35 pKa = 4.42LTPEE39 pKa = 3.87VSGVLRR45 pKa = 11.84SGSGRR50 pKa = 11.84RR51 pKa = 11.84NVNGIKK57 pKa = 10.26SKK59 pKa = 10.95NKK61 pKa = 10.19SYY63 pKa = 11.35DD64 pKa = 3.44FNQVFSDD71 pKa = 3.28IALTLTSAISNPLVLLTIALSVGVITTHH99 pKa = 6.9NFPQNKK105 pKa = 9.01GFIYY109 pKa = 9.2DD110 pKa = 3.87TFRR113 pKa = 11.84EE114 pKa = 4.23KK115 pKa = 11.15NDD117 pKa = 3.94TISRR121 pKa = 11.84WVVDD125 pKa = 3.43NGQKK129 pKa = 10.12VAGFSIFLPAVVDD142 pKa = 3.7SPRR145 pKa = 11.84NFRR148 pKa = 11.84SVIALTSFLWVMVIPEE164 pKa = 4.3SSVVEE169 pKa = 4.14YY170 pKa = 10.0FLQALAVHH178 pKa = 6.63TYY180 pKa = 10.2FRR182 pKa = 11.84LKK184 pKa = 9.76TDD186 pKa = 3.24NSRR189 pKa = 11.84LTLLAIIGIAWFLGYY204 pKa = 8.88FHH206 pKa = 7.6IPTTKK211 pKa = 10.49
Molecular weight: 23.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3187 |
211 |
2550 |
1062.3 |
121.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.801 ± 0.372 | 2.604 ± 0.595 |
6.15 ± 0.71 | 4.518 ± 0.791 |
6.056 ± 1.135 | 3.483 ± 0.502 |
3.044 ± 0.334 | 6.37 ± 0.358 |
5.083 ± 0.76 | 9.162 ± 1.171 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.008 ± 0.627 | 5.742 ± 0.305 |
3.671 ± 0.217 | 2.479 ± 0.229 |
5.24 ± 0.555 | 9.727 ± 1.222 |
5.491 ± 0.337 | 8.033 ± 0.395 |
0.565 ± 0.171 | 5.773 ± 0.679 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
