
Magnaporthe oryzae polymycovirus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
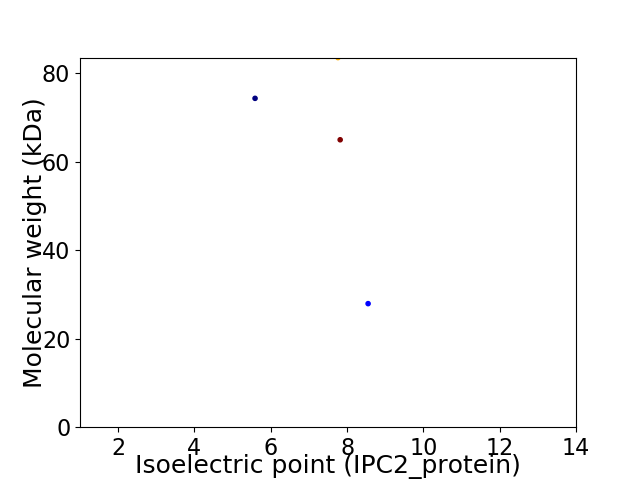
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
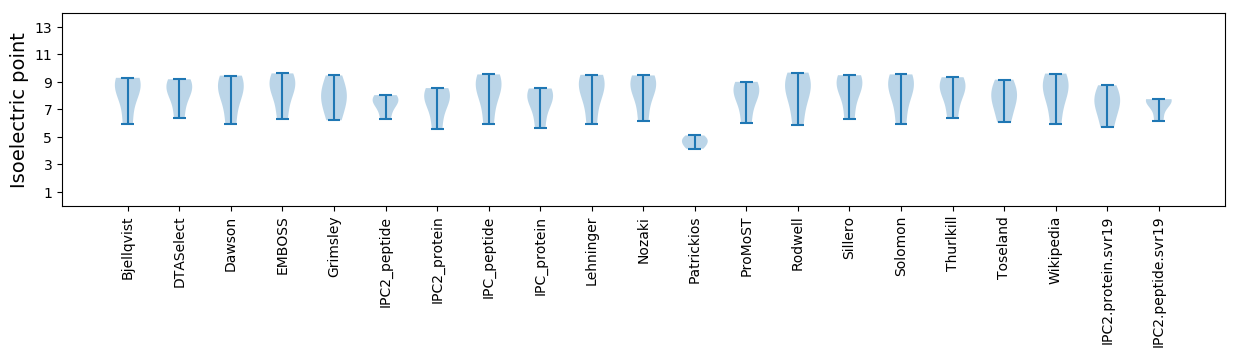
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A410TEN7|A0A410TEN7_9VIRU Uncharacterized protein OS=Magnaporthe oryzae polymycovirus 1 OX=2509266 GN=ORF2a PE=2 SV=1
MM1 pKa = 7.93ADD3 pKa = 3.6LTRR6 pKa = 11.84LRR8 pKa = 11.84SILLDD13 pKa = 3.47TATPAAQIGMALYY26 pKa = 10.92HH27 pKa = 6.42MFPAPDD33 pKa = 3.46ASALLDD39 pKa = 3.78SQSVRR44 pKa = 11.84QAVLWLSSAPVALYY58 pKa = 10.75GGVVGAAGAQPSDD71 pKa = 3.68FARR74 pKa = 11.84LLDD77 pKa = 4.21KK78 pKa = 11.31LDD80 pKa = 4.53DD81 pKa = 3.83STAARR86 pKa = 11.84AATIAHH92 pKa = 6.2AMVARR97 pKa = 11.84DD98 pKa = 4.08RR99 pKa = 11.84EE100 pKa = 3.84WDD102 pKa = 3.42RR103 pKa = 11.84AVRR106 pKa = 11.84DD107 pKa = 4.51GYY109 pKa = 8.06QTQVSVHH116 pKa = 6.11GPGSARR122 pKa = 11.84VDD124 pKa = 3.17AQTSVEE130 pKa = 4.34FAALDD135 pKa = 3.74ALTDD139 pKa = 3.5AGYY142 pKa = 11.05AGIDD146 pKa = 3.51DD147 pKa = 4.44AVAAFADD154 pKa = 3.92LRR156 pKa = 11.84RR157 pKa = 11.84VVAKK161 pKa = 6.94STCVIRR167 pKa = 11.84STQLARR173 pKa = 11.84SRR175 pKa = 11.84HH176 pKa = 4.42AVIARR181 pKa = 11.84FPLSDD186 pKa = 3.21GTVRR190 pKa = 11.84LVFTPWCLSRR200 pKa = 11.84RR201 pKa = 11.84SALAVAGLLVSDD213 pKa = 3.74VRR215 pKa = 11.84RR216 pKa = 11.84GVLHH220 pKa = 6.62DD221 pKa = 3.8RR222 pKa = 11.84VVKK225 pKa = 10.47RR226 pKa = 11.84ALTVTTFAARR236 pKa = 11.84TSLQAVRR243 pKa = 11.84DD244 pKa = 3.85LTGPALALLTRR255 pKa = 11.84YY256 pKa = 10.09AYY258 pKa = 10.53DD259 pKa = 3.81PVRR262 pKa = 11.84MALVGDD268 pKa = 4.73DD269 pKa = 3.94GVEE272 pKa = 3.94QQTVGSRR279 pKa = 11.84LPHH282 pKa = 4.56VTVAFAAIFSSGSADD297 pKa = 3.55PLIALARR304 pKa = 11.84ARR306 pKa = 11.84FEE308 pKa = 4.41LANGLPNLNMNASARR323 pKa = 11.84QYY325 pKa = 10.88FEE327 pKa = 4.26YY328 pKa = 11.12AEE330 pKa = 4.8DD331 pKa = 4.61PYY333 pKa = 11.66SDD335 pKa = 3.66VVLFSAALAQARR347 pKa = 11.84QQRR350 pKa = 11.84EE351 pKa = 4.05KK352 pKa = 10.74SEE354 pKa = 3.82HH355 pKa = 3.92GHH357 pKa = 5.68RR358 pKa = 11.84VNKK361 pKa = 10.05GAMLGYY367 pKa = 9.22VARR370 pKa = 11.84GRR372 pKa = 11.84TALDD376 pKa = 3.78RR377 pKa = 11.84NTSACVNRR385 pKa = 11.84IEE387 pKa = 4.32EE388 pKa = 4.47TVVTLRR394 pKa = 11.84ARR396 pKa = 11.84GHH398 pKa = 5.94EE399 pKa = 3.97PSSYY403 pKa = 10.73IMVVEE408 pKa = 4.1WGGAINIATVLASAAVAGIDD428 pKa = 3.73VALDD432 pKa = 3.57VADD435 pKa = 4.75SGIDD439 pKa = 3.51LPGADD444 pKa = 4.22IYY446 pKa = 11.63GDD448 pKa = 3.54NDD450 pKa = 3.88DD451 pKa = 4.92PAHH454 pKa = 6.97HH455 pKa = 6.2YY456 pKa = 10.13QLYY459 pKa = 9.47LARR462 pKa = 11.84ASSMNLPRR470 pKa = 11.84MPTVEE475 pKa = 4.19YY476 pKa = 10.25TKK478 pKa = 10.1GTPLVVKK485 pKa = 10.55LEE487 pKa = 4.43SVLSAVKK494 pKa = 10.22AAGDD498 pKa = 3.25KK499 pKa = 10.61RR500 pKa = 11.84LVYY503 pKa = 10.06VHH505 pKa = 6.69GGIARR510 pKa = 11.84EE511 pKa = 3.9RR512 pKa = 11.84QVPISTAIDD521 pKa = 3.45CNNRR525 pKa = 11.84LDD527 pKa = 4.16ALSHH531 pKa = 6.22VIADD535 pKa = 4.65PPPLLYY541 pKa = 10.25TSEE544 pKa = 4.0ILLPPFCVHH553 pKa = 7.22SYY555 pKa = 10.27QCTAEE560 pKa = 4.32ALLEE564 pKa = 4.21TGGLVDD570 pKa = 5.0DD571 pKa = 4.95TCDD574 pKa = 3.17QCEE577 pKa = 4.17LVSRR581 pKa = 11.84ACAVLSSILDD591 pKa = 3.43RR592 pKa = 11.84PGIRR596 pKa = 11.84LVKK599 pKa = 9.74ARR601 pKa = 11.84SMYY604 pKa = 10.53AHH606 pKa = 6.86NSHH609 pKa = 6.88FAVEE613 pKa = 4.16VFPGEE618 pKa = 3.92VDD620 pKa = 3.88LSHH623 pKa = 7.3DD624 pKa = 3.69TATTTDD630 pKa = 2.9ACVAANVLRR639 pKa = 11.84NHH641 pKa = 7.45TYY643 pKa = 7.92NTPPPEE649 pKa = 5.29GDD651 pKa = 2.97ASKK654 pKa = 10.99DD655 pKa = 3.19ITSPEE660 pKa = 4.05LMEE663 pKa = 3.98LTRR666 pKa = 11.84PIIEE670 pKa = 4.24MVSAAMSGGPVAKK683 pKa = 10.49VSAEE687 pKa = 3.98RR688 pKa = 11.84ADD690 pKa = 3.55QVAASIAA697 pKa = 3.4
MM1 pKa = 7.93ADD3 pKa = 3.6LTRR6 pKa = 11.84LRR8 pKa = 11.84SILLDD13 pKa = 3.47TATPAAQIGMALYY26 pKa = 10.92HH27 pKa = 6.42MFPAPDD33 pKa = 3.46ASALLDD39 pKa = 3.78SQSVRR44 pKa = 11.84QAVLWLSSAPVALYY58 pKa = 10.75GGVVGAAGAQPSDD71 pKa = 3.68FARR74 pKa = 11.84LLDD77 pKa = 4.21KK78 pKa = 11.31LDD80 pKa = 4.53DD81 pKa = 3.83STAARR86 pKa = 11.84AATIAHH92 pKa = 6.2AMVARR97 pKa = 11.84DD98 pKa = 4.08RR99 pKa = 11.84EE100 pKa = 3.84WDD102 pKa = 3.42RR103 pKa = 11.84AVRR106 pKa = 11.84DD107 pKa = 4.51GYY109 pKa = 8.06QTQVSVHH116 pKa = 6.11GPGSARR122 pKa = 11.84VDD124 pKa = 3.17AQTSVEE130 pKa = 4.34FAALDD135 pKa = 3.74ALTDD139 pKa = 3.5AGYY142 pKa = 11.05AGIDD146 pKa = 3.51DD147 pKa = 4.44AVAAFADD154 pKa = 3.92LRR156 pKa = 11.84RR157 pKa = 11.84VVAKK161 pKa = 6.94STCVIRR167 pKa = 11.84STQLARR173 pKa = 11.84SRR175 pKa = 11.84HH176 pKa = 4.42AVIARR181 pKa = 11.84FPLSDD186 pKa = 3.21GTVRR190 pKa = 11.84LVFTPWCLSRR200 pKa = 11.84RR201 pKa = 11.84SALAVAGLLVSDD213 pKa = 3.74VRR215 pKa = 11.84RR216 pKa = 11.84GVLHH220 pKa = 6.62DD221 pKa = 3.8RR222 pKa = 11.84VVKK225 pKa = 10.47RR226 pKa = 11.84ALTVTTFAARR236 pKa = 11.84TSLQAVRR243 pKa = 11.84DD244 pKa = 3.85LTGPALALLTRR255 pKa = 11.84YY256 pKa = 10.09AYY258 pKa = 10.53DD259 pKa = 3.81PVRR262 pKa = 11.84MALVGDD268 pKa = 4.73DD269 pKa = 3.94GVEE272 pKa = 3.94QQTVGSRR279 pKa = 11.84LPHH282 pKa = 4.56VTVAFAAIFSSGSADD297 pKa = 3.55PLIALARR304 pKa = 11.84ARR306 pKa = 11.84FEE308 pKa = 4.41LANGLPNLNMNASARR323 pKa = 11.84QYY325 pKa = 10.88FEE327 pKa = 4.26YY328 pKa = 11.12AEE330 pKa = 4.8DD331 pKa = 4.61PYY333 pKa = 11.66SDD335 pKa = 3.66VVLFSAALAQARR347 pKa = 11.84QQRR350 pKa = 11.84EE351 pKa = 4.05KK352 pKa = 10.74SEE354 pKa = 3.82HH355 pKa = 3.92GHH357 pKa = 5.68RR358 pKa = 11.84VNKK361 pKa = 10.05GAMLGYY367 pKa = 9.22VARR370 pKa = 11.84GRR372 pKa = 11.84TALDD376 pKa = 3.78RR377 pKa = 11.84NTSACVNRR385 pKa = 11.84IEE387 pKa = 4.32EE388 pKa = 4.47TVVTLRR394 pKa = 11.84ARR396 pKa = 11.84GHH398 pKa = 5.94EE399 pKa = 3.97PSSYY403 pKa = 10.73IMVVEE408 pKa = 4.1WGGAINIATVLASAAVAGIDD428 pKa = 3.73VALDD432 pKa = 3.57VADD435 pKa = 4.75SGIDD439 pKa = 3.51LPGADD444 pKa = 4.22IYY446 pKa = 11.63GDD448 pKa = 3.54NDD450 pKa = 3.88DD451 pKa = 4.92PAHH454 pKa = 6.97HH455 pKa = 6.2YY456 pKa = 10.13QLYY459 pKa = 9.47LARR462 pKa = 11.84ASSMNLPRR470 pKa = 11.84MPTVEE475 pKa = 4.19YY476 pKa = 10.25TKK478 pKa = 10.1GTPLVVKK485 pKa = 10.55LEE487 pKa = 4.43SVLSAVKK494 pKa = 10.22AAGDD498 pKa = 3.25KK499 pKa = 10.61RR500 pKa = 11.84LVYY503 pKa = 10.06VHH505 pKa = 6.69GGIARR510 pKa = 11.84EE511 pKa = 3.9RR512 pKa = 11.84QVPISTAIDD521 pKa = 3.45CNNRR525 pKa = 11.84LDD527 pKa = 4.16ALSHH531 pKa = 6.22VIADD535 pKa = 4.65PPPLLYY541 pKa = 10.25TSEE544 pKa = 4.0ILLPPFCVHH553 pKa = 7.22SYY555 pKa = 10.27QCTAEE560 pKa = 4.32ALLEE564 pKa = 4.21TGGLVDD570 pKa = 5.0DD571 pKa = 4.95TCDD574 pKa = 3.17QCEE577 pKa = 4.17LVSRR581 pKa = 11.84ACAVLSSILDD591 pKa = 3.43RR592 pKa = 11.84PGIRR596 pKa = 11.84LVKK599 pKa = 9.74ARR601 pKa = 11.84SMYY604 pKa = 10.53AHH606 pKa = 6.86NSHH609 pKa = 6.88FAVEE613 pKa = 4.16VFPGEE618 pKa = 3.92VDD620 pKa = 3.88LSHH623 pKa = 7.3DD624 pKa = 3.69TATTTDD630 pKa = 2.9ACVAANVLRR639 pKa = 11.84NHH641 pKa = 7.45TYY643 pKa = 7.92NTPPPEE649 pKa = 5.29GDD651 pKa = 2.97ASKK654 pKa = 10.99DD655 pKa = 3.19ITSPEE660 pKa = 4.05LMEE663 pKa = 3.98LTRR666 pKa = 11.84PIIEE670 pKa = 4.24MVSAAMSGGPVAKK683 pKa = 10.49VSAEE687 pKa = 3.98RR688 pKa = 11.84ADD690 pKa = 3.55QVAASIAA697 pKa = 3.4
Molecular weight: 74.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A410TEM9|A0A410TEM9_9VIRU Uncharacterized protein OS=Magnaporthe oryzae polymycovirus 1 OX=2509266 GN=ORF3a PE=2 SV=1
MM1 pKa = 7.52SLNDD5 pKa = 3.69SVSAATAKK13 pKa = 10.7SLTQHH18 pKa = 5.72SPEE21 pKa = 4.08VFTTFARR28 pKa = 11.84LVTLGFSPSEE38 pKa = 3.58LRR40 pKa = 11.84AALVRR45 pKa = 11.84VANGEE50 pKa = 4.15EE51 pKa = 4.36VPLPGNLGRR60 pKa = 11.84PRR62 pKa = 11.84PLTINAWSFCVDD74 pKa = 3.27RR75 pKa = 11.84GQYY78 pKa = 10.8ADD80 pKa = 3.95TYY82 pKa = 11.33GLDD85 pKa = 3.5DD86 pKa = 4.45SEE88 pKa = 4.79AGRR91 pKa = 11.84LKK93 pKa = 10.94GVLRR97 pKa = 11.84ANADD101 pKa = 3.59DD102 pKa = 4.47GVAEE106 pKa = 4.12ITKK109 pKa = 10.53LVTDD113 pKa = 4.4KK114 pKa = 11.35LRR116 pKa = 11.84ISGSARR122 pKa = 11.84AVHH125 pKa = 5.01VTKK128 pKa = 10.64SGMPQSVVTGSGLKK142 pKa = 9.78PGKK145 pKa = 10.1QGGADD150 pKa = 3.79LKK152 pKa = 10.22TVILTNPALFGMYY165 pKa = 9.47TFVPSDD171 pKa = 3.46SGVPGPLRR179 pKa = 11.84YY180 pKa = 9.31RR181 pKa = 11.84CRR183 pKa = 11.84IGSGLYY189 pKa = 10.43ACFPSKK195 pKa = 10.47KK196 pKa = 10.0GAIDD200 pKa = 3.2VARR203 pKa = 11.84ICRR206 pKa = 11.84VHH208 pKa = 5.87GRR210 pKa = 11.84NHH212 pKa = 5.35EE213 pKa = 4.56AIAGRR218 pKa = 11.84VYY220 pKa = 10.45FIRR223 pKa = 11.84EE224 pKa = 3.71GKK226 pKa = 10.55APLTGADD233 pKa = 3.12IPEE236 pKa = 4.15KK237 pKa = 11.01VKK239 pKa = 10.83FDD241 pKa = 3.4GKK243 pKa = 10.49LRR245 pKa = 11.84PDD247 pKa = 3.73EE248 pKa = 4.67PVPADD253 pKa = 3.64PVNSPTTEE261 pKa = 3.92VNN263 pKa = 3.08
MM1 pKa = 7.52SLNDD5 pKa = 3.69SVSAATAKK13 pKa = 10.7SLTQHH18 pKa = 5.72SPEE21 pKa = 4.08VFTTFARR28 pKa = 11.84LVTLGFSPSEE38 pKa = 3.58LRR40 pKa = 11.84AALVRR45 pKa = 11.84VANGEE50 pKa = 4.15EE51 pKa = 4.36VPLPGNLGRR60 pKa = 11.84PRR62 pKa = 11.84PLTINAWSFCVDD74 pKa = 3.27RR75 pKa = 11.84GQYY78 pKa = 10.8ADD80 pKa = 3.95TYY82 pKa = 11.33GLDD85 pKa = 3.5DD86 pKa = 4.45SEE88 pKa = 4.79AGRR91 pKa = 11.84LKK93 pKa = 10.94GVLRR97 pKa = 11.84ANADD101 pKa = 3.59DD102 pKa = 4.47GVAEE106 pKa = 4.12ITKK109 pKa = 10.53LVTDD113 pKa = 4.4KK114 pKa = 11.35LRR116 pKa = 11.84ISGSARR122 pKa = 11.84AVHH125 pKa = 5.01VTKK128 pKa = 10.64SGMPQSVVTGSGLKK142 pKa = 9.78PGKK145 pKa = 10.1QGGADD150 pKa = 3.79LKK152 pKa = 10.22TVILTNPALFGMYY165 pKa = 9.47TFVPSDD171 pKa = 3.46SGVPGPLRR179 pKa = 11.84YY180 pKa = 9.31RR181 pKa = 11.84CRR183 pKa = 11.84IGSGLYY189 pKa = 10.43ACFPSKK195 pKa = 10.47KK196 pKa = 10.0GAIDD200 pKa = 3.2VARR203 pKa = 11.84ICRR206 pKa = 11.84VHH208 pKa = 5.87GRR210 pKa = 11.84NHH212 pKa = 5.35EE213 pKa = 4.56AIAGRR218 pKa = 11.84VYY220 pKa = 10.45FIRR223 pKa = 11.84EE224 pKa = 3.71GKK226 pKa = 10.55APLTGADD233 pKa = 3.12IPEE236 pKa = 4.15KK237 pKa = 11.01VKK239 pKa = 10.83FDD241 pKa = 3.4GKK243 pKa = 10.49LRR245 pKa = 11.84PDD247 pKa = 3.73EE248 pKa = 4.67PVPADD253 pKa = 3.64PVNSPTTEE261 pKa = 3.92VNN263 pKa = 3.08
Molecular weight: 27.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2338 |
263 |
766 |
584.5 |
62.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.19 ± 1.199 | 0.941 ± 0.212 |
6.373 ± 0.289 | 4.063 ± 0.369 |
2.866 ± 0.36 | 7.613 ± 0.776 |
2.524 ± 0.173 | 3.678 ± 0.058 |
2.823 ± 0.518 | 9.196 ± 0.472 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.139 ± 0.203 | 2.395 ± 0.182 |
6.373 ± 0.559 | 2.31 ± 0.217 |
7.742 ± 0.238 | 7.699 ± 0.153 |
6.33 ± 0.166 | 9.324 ± 0.811 |
0.642 ± 0.059 | 2.78 ± 0.345 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
