
Murine coronavirus (strain 2) (MHV-2) (Murine hepatitis virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Embecovirus; Murine coronavirus; Murine hepatitis virus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
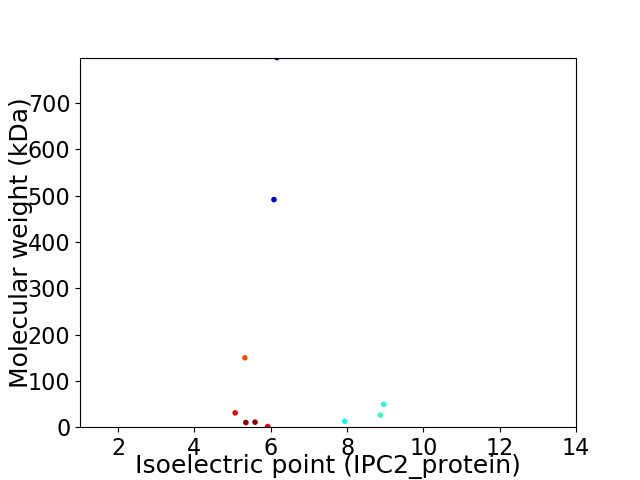
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
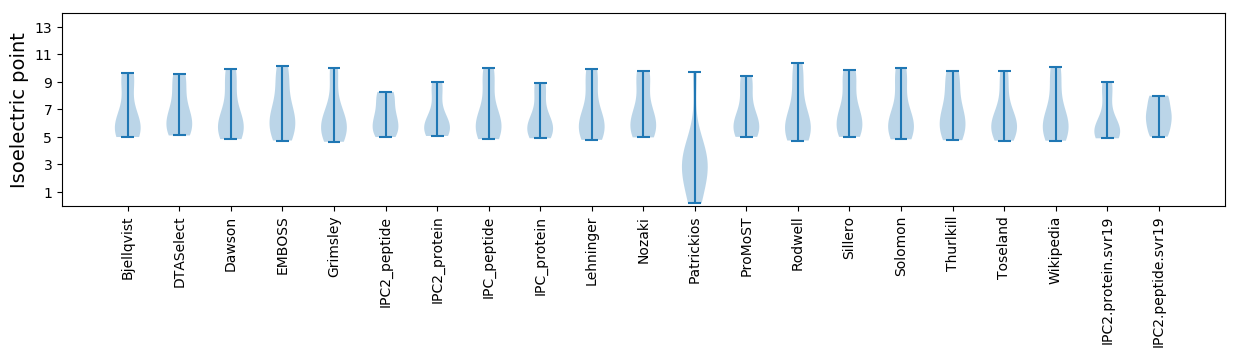
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9PYA1|Q9PYA1_CVM2 Non-structural protein 2a OS=Murine coronavirus (strain 2) OX=76344 PE=2 SV=1
MM1 pKa = 7.67AAMQAFADD9 pKa = 4.26KK10 pKa = 9.98PNHH13 pKa = 6.87FINFPLAQFSGVMGKK28 pKa = 7.96YY29 pKa = 10.3LKK31 pKa = 10.59LQSQLVEE38 pKa = 4.08MGLDD42 pKa = 3.76CKK44 pKa = 10.5LQKK47 pKa = 10.45APHH50 pKa = 5.8VSITMLDD57 pKa = 3.25IRR59 pKa = 11.84ADD61 pKa = 3.31QYY63 pKa = 11.5KK64 pKa = 9.51QVEE67 pKa = 4.17FAIQEE72 pKa = 4.43VIDD75 pKa = 5.23DD76 pKa = 3.74MAAYY80 pKa = 9.87EE81 pKa = 4.12GDD83 pKa = 3.31IVFDD87 pKa = 3.88NPHH90 pKa = 5.62MLGRR94 pKa = 11.84CLVLDD99 pKa = 3.42VKK101 pKa = 10.99GFEE104 pKa = 4.45EE105 pKa = 4.03LHH107 pKa = 6.06EE108 pKa = 5.39DD109 pKa = 2.99IVEE112 pKa = 4.05ILRR115 pKa = 11.84KK116 pKa = 9.49RR117 pKa = 11.84GCIADD122 pKa = 4.37QSRR125 pKa = 11.84QWIPHH130 pKa = 4.93CTVAQFEE137 pKa = 4.44EE138 pKa = 4.34EE139 pKa = 4.24KK140 pKa = 10.66EE141 pKa = 4.02INEE144 pKa = 3.8LQFYY148 pKa = 10.15YY149 pKa = 10.98KK150 pKa = 10.7LPFYY154 pKa = 10.68LKK156 pKa = 10.34HH157 pKa = 6.46NNILTDD163 pKa = 3.31ARR165 pKa = 11.84LEE167 pKa = 4.03LVKK170 pKa = 10.53IGSSKK175 pKa = 9.95TDD177 pKa = 2.85GFYY180 pKa = 10.87CSEE183 pKa = 3.82LSIWCGEE190 pKa = 4.08RR191 pKa = 11.84LCYY194 pKa = 10.09KK195 pKa = 10.34PPTPKK200 pKa = 10.32FSDD203 pKa = 2.77IFGYY207 pKa = 10.69CCIDD211 pKa = 4.07KK212 pKa = 10.67IRR214 pKa = 11.84VDD216 pKa = 4.68LEE218 pKa = 4.38IGDD221 pKa = 4.36LPEE224 pKa = 6.18DD225 pKa = 4.99DD226 pKa = 3.96EE227 pKa = 4.58EE228 pKa = 5.02AWAEE232 pKa = 3.96LSYY235 pKa = 10.57HH236 pKa = 4.98YY237 pKa = 10.55QRR239 pKa = 11.84NTYY242 pKa = 8.74FFRR245 pKa = 11.84YY246 pKa = 9.06VHH248 pKa = 7.01DD249 pKa = 3.57NSIYY253 pKa = 9.8FRR255 pKa = 11.84TVCRR259 pKa = 11.84MKK261 pKa = 11.07GCMCC265 pKa = 5.37
MM1 pKa = 7.67AAMQAFADD9 pKa = 4.26KK10 pKa = 9.98PNHH13 pKa = 6.87FINFPLAQFSGVMGKK28 pKa = 7.96YY29 pKa = 10.3LKK31 pKa = 10.59LQSQLVEE38 pKa = 4.08MGLDD42 pKa = 3.76CKK44 pKa = 10.5LQKK47 pKa = 10.45APHH50 pKa = 5.8VSITMLDD57 pKa = 3.25IRR59 pKa = 11.84ADD61 pKa = 3.31QYY63 pKa = 11.5KK64 pKa = 9.51QVEE67 pKa = 4.17FAIQEE72 pKa = 4.43VIDD75 pKa = 5.23DD76 pKa = 3.74MAAYY80 pKa = 9.87EE81 pKa = 4.12GDD83 pKa = 3.31IVFDD87 pKa = 3.88NPHH90 pKa = 5.62MLGRR94 pKa = 11.84CLVLDD99 pKa = 3.42VKK101 pKa = 10.99GFEE104 pKa = 4.45EE105 pKa = 4.03LHH107 pKa = 6.06EE108 pKa = 5.39DD109 pKa = 2.99IVEE112 pKa = 4.05ILRR115 pKa = 11.84KK116 pKa = 9.49RR117 pKa = 11.84GCIADD122 pKa = 4.37QSRR125 pKa = 11.84QWIPHH130 pKa = 4.93CTVAQFEE137 pKa = 4.44EE138 pKa = 4.34EE139 pKa = 4.24KK140 pKa = 10.66EE141 pKa = 4.02INEE144 pKa = 3.8LQFYY148 pKa = 10.15YY149 pKa = 10.98KK150 pKa = 10.7LPFYY154 pKa = 10.68LKK156 pKa = 10.34HH157 pKa = 6.46NNILTDD163 pKa = 3.31ARR165 pKa = 11.84LEE167 pKa = 4.03LVKK170 pKa = 10.53IGSSKK175 pKa = 9.95TDD177 pKa = 2.85GFYY180 pKa = 10.87CSEE183 pKa = 3.82LSIWCGEE190 pKa = 4.08RR191 pKa = 11.84LCYY194 pKa = 10.09KK195 pKa = 10.34PPTPKK200 pKa = 10.32FSDD203 pKa = 2.77IFGYY207 pKa = 10.69CCIDD211 pKa = 4.07KK212 pKa = 10.67IRR214 pKa = 11.84VDD216 pKa = 4.68LEE218 pKa = 4.38IGDD221 pKa = 4.36LPEE224 pKa = 6.18DD225 pKa = 4.99DD226 pKa = 3.96EE227 pKa = 4.58EE228 pKa = 5.02AWAEE232 pKa = 3.96LSYY235 pKa = 10.57HH236 pKa = 4.98YY237 pKa = 10.55QRR239 pKa = 11.84NTYY242 pKa = 8.74FFRR245 pKa = 11.84YY246 pKa = 9.06VHH248 pKa = 7.01DD249 pKa = 3.57NSIYY253 pKa = 9.8FRR255 pKa = 11.84TVCRR259 pKa = 11.84MKK261 pKa = 11.07GCMCC265 pKa = 5.37
Molecular weight: 30.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9PY98|Q9PY98_CVM2 Non-structural protein OS=Murine coronavirus (strain 2) OX=76344 PE=2 SV=1
MM1 pKa = 7.35SFVPGQEE8 pKa = 4.06NAGSRR13 pKa = 11.84SSSGNRR19 pKa = 11.84AGNGILKK26 pKa = 8.59KK27 pKa = 7.37TTWADD32 pKa = 3.05QTEE35 pKa = 4.24RR36 pKa = 11.84GNRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84NHH44 pKa = 6.79PKK46 pKa = 8.46QTATTQPNAGSVVPHH61 pKa = 5.92YY62 pKa = 10.92SWFSGITQFQKK73 pKa = 10.82GKK75 pKa = 9.99EE76 pKa = 3.93FQFAQGQGVPIASGIPASEE95 pKa = 4.15QKK97 pKa = 10.87GYY99 pKa = 9.69WYY101 pKa = 10.34RR102 pKa = 11.84HH103 pKa = 4.36NRR105 pKa = 11.84RR106 pKa = 11.84SFKK109 pKa = 10.77TPDD112 pKa = 3.54GQHH115 pKa = 5.87KK116 pKa = 10.03QLLPRR121 pKa = 11.84WYY123 pKa = 9.79FYY125 pKa = 11.56YY126 pKa = 10.78LGTGPHH132 pKa = 6.74AGAEE136 pKa = 4.09YY137 pKa = 10.75GDD139 pKa = 5.04DD140 pKa = 3.44IEE142 pKa = 4.76GVVWVASQQADD153 pKa = 3.61TKK155 pKa = 8.29TTADD159 pKa = 3.4VVEE162 pKa = 4.91RR163 pKa = 11.84DD164 pKa = 3.89PSSHH168 pKa = 5.75EE169 pKa = 4.53AIPTRR174 pKa = 11.84FAPGTVLPQGFYY186 pKa = 10.92VEE188 pKa = 4.42GSGRR192 pKa = 11.84SAPASRR198 pKa = 11.84SGSRR202 pKa = 11.84SQSRR206 pKa = 11.84GPNNRR211 pKa = 11.84ARR213 pKa = 11.84SSSNQRR219 pKa = 11.84QPASAVKK226 pKa = 9.82PDD228 pKa = 3.5MAEE231 pKa = 3.97EE232 pKa = 3.7IAALVLAKK240 pKa = 10.33LGKK243 pKa = 10.0DD244 pKa = 2.88AGQPKK249 pKa = 9.61QVTKK253 pKa = 10.71QSAKK257 pKa = 8.91EE258 pKa = 3.89VRR260 pKa = 11.84QKK262 pKa = 10.93ILTKK266 pKa = 9.83PRR268 pKa = 11.84QKK270 pKa = 9.65RR271 pKa = 11.84TPNKK275 pKa = 8.97QCPVQQCFGKK285 pKa = 10.52RR286 pKa = 11.84GPNQNFGGSEE296 pKa = 3.94MLKK299 pKa = 10.6LGTSDD304 pKa = 3.38PQFPILAEE312 pKa = 4.09LAPTPSAFFFGSKK325 pKa = 10.65LEE327 pKa = 4.03LVKK330 pKa = 10.9KK331 pKa = 10.59NSGGADD337 pKa = 3.41EE338 pKa = 4.48PTKK341 pKa = 10.65DD342 pKa = 3.71VYY344 pKa = 10.84EE345 pKa = 4.13LQYY348 pKa = 11.19SGAIRR353 pKa = 11.84FDD355 pKa = 3.3STLPGFEE362 pKa = 4.85TIMKK366 pKa = 10.09VLTEE370 pKa = 4.26NLNAYY375 pKa = 9.23QDD377 pKa = 3.65QAGSVDD383 pKa = 4.3LVSPKK388 pKa = 9.78PPRR391 pKa = 11.84RR392 pKa = 11.84GRR394 pKa = 11.84RR395 pKa = 11.84QAQEE399 pKa = 4.04KK400 pKa = 10.0KK401 pKa = 10.78DD402 pKa = 3.67EE403 pKa = 4.08VDD405 pKa = 3.28NVSVAKK411 pKa = 10.22PKK413 pKa = 10.9SLVQRR418 pKa = 11.84NVSRR422 pKa = 11.84EE423 pKa = 4.02LTPEE427 pKa = 4.02DD428 pKa = 4.12RR429 pKa = 11.84SLLAQILDD437 pKa = 4.15DD438 pKa = 4.33GVVPDD443 pKa = 4.88GLEE446 pKa = 4.2DD447 pKa = 4.13DD448 pKa = 4.85SNVV451 pKa = 3.06
MM1 pKa = 7.35SFVPGQEE8 pKa = 4.06NAGSRR13 pKa = 11.84SSSGNRR19 pKa = 11.84AGNGILKK26 pKa = 8.59KK27 pKa = 7.37TTWADD32 pKa = 3.05QTEE35 pKa = 4.24RR36 pKa = 11.84GNRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84NHH44 pKa = 6.79PKK46 pKa = 8.46QTATTQPNAGSVVPHH61 pKa = 5.92YY62 pKa = 10.92SWFSGITQFQKK73 pKa = 10.82GKK75 pKa = 9.99EE76 pKa = 3.93FQFAQGQGVPIASGIPASEE95 pKa = 4.15QKK97 pKa = 10.87GYY99 pKa = 9.69WYY101 pKa = 10.34RR102 pKa = 11.84HH103 pKa = 4.36NRR105 pKa = 11.84RR106 pKa = 11.84SFKK109 pKa = 10.77TPDD112 pKa = 3.54GQHH115 pKa = 5.87KK116 pKa = 10.03QLLPRR121 pKa = 11.84WYY123 pKa = 9.79FYY125 pKa = 11.56YY126 pKa = 10.78LGTGPHH132 pKa = 6.74AGAEE136 pKa = 4.09YY137 pKa = 10.75GDD139 pKa = 5.04DD140 pKa = 3.44IEE142 pKa = 4.76GVVWVASQQADD153 pKa = 3.61TKK155 pKa = 8.29TTADD159 pKa = 3.4VVEE162 pKa = 4.91RR163 pKa = 11.84DD164 pKa = 3.89PSSHH168 pKa = 5.75EE169 pKa = 4.53AIPTRR174 pKa = 11.84FAPGTVLPQGFYY186 pKa = 10.92VEE188 pKa = 4.42GSGRR192 pKa = 11.84SAPASRR198 pKa = 11.84SGSRR202 pKa = 11.84SQSRR206 pKa = 11.84GPNNRR211 pKa = 11.84ARR213 pKa = 11.84SSSNQRR219 pKa = 11.84QPASAVKK226 pKa = 9.82PDD228 pKa = 3.5MAEE231 pKa = 3.97EE232 pKa = 3.7IAALVLAKK240 pKa = 10.33LGKK243 pKa = 10.0DD244 pKa = 2.88AGQPKK249 pKa = 9.61QVTKK253 pKa = 10.71QSAKK257 pKa = 8.91EE258 pKa = 3.89VRR260 pKa = 11.84QKK262 pKa = 10.93ILTKK266 pKa = 9.83PRR268 pKa = 11.84QKK270 pKa = 9.65RR271 pKa = 11.84TPNKK275 pKa = 8.97QCPVQQCFGKK285 pKa = 10.52RR286 pKa = 11.84GPNQNFGGSEE296 pKa = 3.94MLKK299 pKa = 10.6LGTSDD304 pKa = 3.38PQFPILAEE312 pKa = 4.09LAPTPSAFFFGSKK325 pKa = 10.65LEE327 pKa = 4.03LVKK330 pKa = 10.9KK331 pKa = 10.59NSGGADD337 pKa = 3.41EE338 pKa = 4.48PTKK341 pKa = 10.65DD342 pKa = 3.71VYY344 pKa = 10.84EE345 pKa = 4.13LQYY348 pKa = 11.19SGAIRR353 pKa = 11.84FDD355 pKa = 3.3STLPGFEE362 pKa = 4.85TIMKK366 pKa = 10.09VLTEE370 pKa = 4.26NLNAYY375 pKa = 9.23QDD377 pKa = 3.65QAGSVDD383 pKa = 4.3LVSPKK388 pKa = 9.78PPRR391 pKa = 11.84RR392 pKa = 11.84GRR394 pKa = 11.84RR395 pKa = 11.84QAQEE399 pKa = 4.04KK400 pKa = 10.0KK401 pKa = 10.78DD402 pKa = 3.67EE403 pKa = 4.08VDD405 pKa = 3.28NVSVAKK411 pKa = 10.22PKK413 pKa = 10.9SLVQRR418 pKa = 11.84NVSRR422 pKa = 11.84EE423 pKa = 4.02LTPEE427 pKa = 4.02DD428 pKa = 4.12RR429 pKa = 11.84SLLAQILDD437 pKa = 4.15DD438 pKa = 4.33GVVPDD443 pKa = 4.88GLEE446 pKa = 4.2DD447 pKa = 4.13DD448 pKa = 4.85SNVV451 pKa = 3.06
Molecular weight: 49.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14151 |
14 |
7124 |
1415.1 |
158.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.918 ± 0.289 | 3.837 ± 0.379 |
5.788 ± 0.279 | 4.056 ± 0.231 |
5.455 ± 0.166 | 6.141 ± 0.336 |
1.661 ± 0.104 | 4.261 ± 0.506 |
5.894 ± 0.399 | 9.264 ± 0.311 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.367 ± 0.174 | 4.897 ± 0.542 |
3.547 ± 0.417 | 3.392 ± 0.535 |
3.611 ± 0.373 | 7.151 ± 0.45 |
5.597 ± 0.211 | 10.105 ± 1.076 |
1.336 ± 0.085 | 4.721 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
