
Blackcurrant reversion virus satellite RNA
Taxonomy: Viruses; Satellites; RNA satellites; Single stranded RNA satellites; unclassified Single stranded RNA satellites
Average proteome isoelectric point is 9.33
Get precalculated fractions of proteins
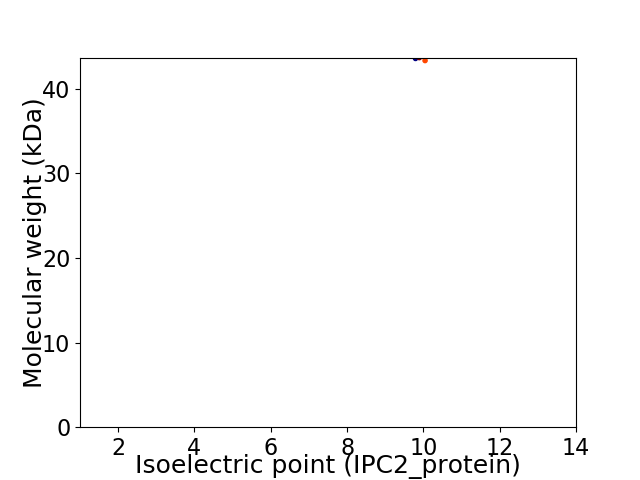
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9QAH2|Q9QAH2_9VIRU Uncharacterized protein OS=Blackcurrant reversion virus satellite RNA OX=99930 PE=4 SV=1
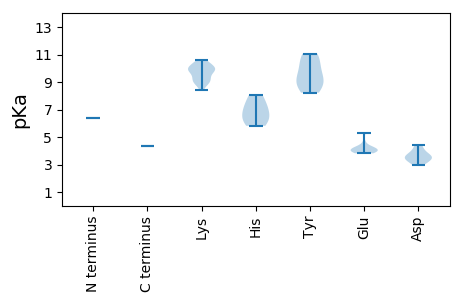
MM1 pKa = 6.36TTSAVHH7 pKa = 7.01DD8 pKa = 3.74SHH10 pKa = 8.06SLRR13 pKa = 11.84KK14 pKa = 9.92GEE16 pKa = 3.85RR17 pKa = 11.84SRR19 pKa = 11.84WTRR22 pKa = 11.84RR23 pKa = 11.84TLSLSVLGPAWRR35 pKa = 11.84KK36 pKa = 8.91MGGTGALRR44 pKa = 11.84GTRR47 pKa = 11.84QRR49 pKa = 11.84GSMWVPVTVVAEE61 pKa = 4.0AVKK64 pKa = 10.43LSRR67 pKa = 11.84RR68 pKa = 11.84AHH70 pKa = 5.82YY71 pKa = 9.38SAPSCANRR79 pKa = 11.84VDD81 pKa = 3.46VRR83 pKa = 11.84AGNGRR88 pKa = 11.84KK89 pKa = 9.27IIAAVVAPATKK100 pKa = 10.0GAKK103 pKa = 9.39KK104 pKa = 10.58APLRR108 pKa = 11.84PSLVSPKK115 pKa = 9.9VVPSSRR121 pKa = 11.84GVSRR125 pKa = 11.84PSGRR129 pKa = 11.84KK130 pKa = 8.45GAYY133 pKa = 8.23SHH135 pKa = 7.38SSGTIDD141 pKa = 3.52NRR143 pKa = 11.84SYY145 pKa = 11.06KK146 pKa = 9.63QALVGNASVKK156 pKa = 8.88EE157 pKa = 4.25TAAKK161 pKa = 9.39PAGEE165 pKa = 3.91FVAAKK170 pKa = 9.99KK171 pKa = 9.76VARR174 pKa = 11.84KK175 pKa = 8.94AATSLPQKK183 pKa = 10.42LKK185 pKa = 9.76TGNKK189 pKa = 9.25FSPLVGFARR198 pKa = 11.84AEE200 pKa = 4.11CSRR203 pKa = 11.84GFNTTTPVKK212 pKa = 9.07RR213 pKa = 11.84TYY215 pKa = 10.23PEE217 pKa = 3.95EE218 pKa = 4.02VVQTGVKK225 pKa = 9.85YY226 pKa = 9.44VSTRR230 pKa = 11.84AGHH233 pKa = 6.51QRR235 pKa = 11.84GCYY238 pKa = 8.81NAASYY243 pKa = 10.66LVEE246 pKa = 5.26FDD248 pKa = 4.4RR249 pKa = 11.84ACCNQVVEE257 pKa = 4.25QPIFEE262 pKa = 4.33KK263 pKa = 10.23RR264 pKa = 11.84TVEE267 pKa = 3.9RR268 pKa = 11.84MVVDD272 pKa = 3.65VEE274 pKa = 4.64GEE276 pKa = 4.13TPRR279 pKa = 11.84AEE281 pKa = 4.15MLKK284 pKa = 10.26SIPPRR289 pKa = 11.84ATSTTLVPVGVEE301 pKa = 3.82SLNWGNAPEE310 pKa = 4.27MAPATAKK317 pKa = 10.17RR318 pKa = 11.84PPWFGQRR325 pKa = 11.84GTTLKK330 pKa = 9.94RR331 pKa = 11.84DD332 pKa = 2.98KK333 pKa = 9.03MTRR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 4.03LSRR341 pKa = 11.84LLSALRR347 pKa = 11.84LARR350 pKa = 11.84TTVNRR355 pKa = 11.84SCSLGEE361 pKa = 3.85AFVWAWTTLGFSGAAPIPSLKK382 pKa = 9.96GWEE385 pKa = 4.11IDD387 pKa = 4.03RR388 pKa = 11.84PSPDD392 pKa = 3.38DD393 pKa = 3.23TAAPLHH399 pKa = 6.23TYY401 pKa = 8.74VV402 pKa = 4.36
MM1 pKa = 6.36TTSAVHH7 pKa = 7.01DD8 pKa = 3.74SHH10 pKa = 8.06SLRR13 pKa = 11.84KK14 pKa = 9.92GEE16 pKa = 3.85RR17 pKa = 11.84SRR19 pKa = 11.84WTRR22 pKa = 11.84RR23 pKa = 11.84TLSLSVLGPAWRR35 pKa = 11.84KK36 pKa = 8.91MGGTGALRR44 pKa = 11.84GTRR47 pKa = 11.84QRR49 pKa = 11.84GSMWVPVTVVAEE61 pKa = 4.0AVKK64 pKa = 10.43LSRR67 pKa = 11.84RR68 pKa = 11.84AHH70 pKa = 5.82YY71 pKa = 9.38SAPSCANRR79 pKa = 11.84VDD81 pKa = 3.46VRR83 pKa = 11.84AGNGRR88 pKa = 11.84KK89 pKa = 9.27IIAAVVAPATKK100 pKa = 10.0GAKK103 pKa = 9.39KK104 pKa = 10.58APLRR108 pKa = 11.84PSLVSPKK115 pKa = 9.9VVPSSRR121 pKa = 11.84GVSRR125 pKa = 11.84PSGRR129 pKa = 11.84KK130 pKa = 8.45GAYY133 pKa = 8.23SHH135 pKa = 7.38SSGTIDD141 pKa = 3.52NRR143 pKa = 11.84SYY145 pKa = 11.06KK146 pKa = 9.63QALVGNASVKK156 pKa = 8.88EE157 pKa = 4.25TAAKK161 pKa = 9.39PAGEE165 pKa = 3.91FVAAKK170 pKa = 9.99KK171 pKa = 9.76VARR174 pKa = 11.84KK175 pKa = 8.94AATSLPQKK183 pKa = 10.42LKK185 pKa = 9.76TGNKK189 pKa = 9.25FSPLVGFARR198 pKa = 11.84AEE200 pKa = 4.11CSRR203 pKa = 11.84GFNTTTPVKK212 pKa = 9.07RR213 pKa = 11.84TYY215 pKa = 10.23PEE217 pKa = 3.95EE218 pKa = 4.02VVQTGVKK225 pKa = 9.85YY226 pKa = 9.44VSTRR230 pKa = 11.84AGHH233 pKa = 6.51QRR235 pKa = 11.84GCYY238 pKa = 8.81NAASYY243 pKa = 10.66LVEE246 pKa = 5.26FDD248 pKa = 4.4RR249 pKa = 11.84ACCNQVVEE257 pKa = 4.25QPIFEE262 pKa = 4.33KK263 pKa = 10.23RR264 pKa = 11.84TVEE267 pKa = 3.9RR268 pKa = 11.84MVVDD272 pKa = 3.65VEE274 pKa = 4.64GEE276 pKa = 4.13TPRR279 pKa = 11.84AEE281 pKa = 4.15MLKK284 pKa = 10.26SIPPRR289 pKa = 11.84ATSTTLVPVGVEE301 pKa = 3.82SLNWGNAPEE310 pKa = 4.27MAPATAKK317 pKa = 10.17RR318 pKa = 11.84PPWFGQRR325 pKa = 11.84GTTLKK330 pKa = 9.94RR331 pKa = 11.84DD332 pKa = 2.98KK333 pKa = 9.03MTRR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 4.03LSRR341 pKa = 11.84LLSALRR347 pKa = 11.84LARR350 pKa = 11.84TTVNRR355 pKa = 11.84SCSLGEE361 pKa = 3.85AFVWAWTTLGFSGAAPIPSLKK382 pKa = 9.96GWEE385 pKa = 4.11IDD387 pKa = 4.03RR388 pKa = 11.84PSPDD392 pKa = 3.38DD393 pKa = 3.23TAAPLHH399 pKa = 6.23TYY401 pKa = 8.74VV402 pKa = 4.36
Molecular weight: 43.58 kDa
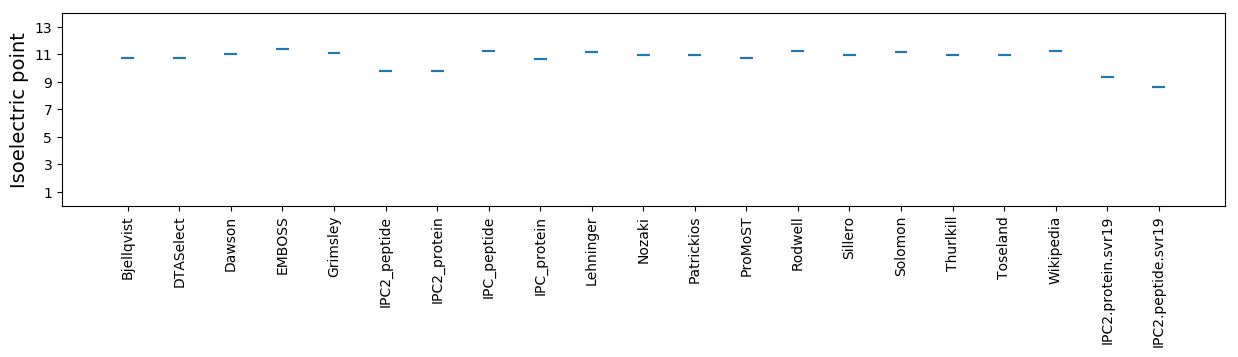
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QAH2|Q9QAH2_9VIRU Uncharacterized protein OS=Blackcurrant reversion virus satellite RNA OX=99930 PE=4 SV=1
MM1 pKa = 6.36TTSAVHH7 pKa = 7.01DD8 pKa = 3.74SHH10 pKa = 8.06SLRR13 pKa = 11.84KK14 pKa = 9.92GEE16 pKa = 3.85RR17 pKa = 11.84SRR19 pKa = 11.84WTRR22 pKa = 11.84RR23 pKa = 11.84TLSLSVLGPAWRR35 pKa = 11.84KK36 pKa = 8.91MGGTGALRR44 pKa = 11.84GTRR47 pKa = 11.84QRR49 pKa = 11.84GSMWVPVTVVAEE61 pKa = 4.0AVKK64 pKa = 10.43LSRR67 pKa = 11.84RR68 pKa = 11.84AHH70 pKa = 5.82YY71 pKa = 9.38SAPSCANRR79 pKa = 11.84VDD81 pKa = 3.46VRR83 pKa = 11.84AGNGRR88 pKa = 11.84KK89 pKa = 9.27IIAAVVAPATKK100 pKa = 10.0GAKK103 pKa = 9.39KK104 pKa = 10.58APLRR108 pKa = 11.84PSLVSPKK115 pKa = 9.9VVPSSRR121 pKa = 11.84GVSRR125 pKa = 11.84PSGRR129 pKa = 11.84KK130 pKa = 8.45GAYY133 pKa = 8.23SHH135 pKa = 7.38SSGTIDD141 pKa = 3.52NRR143 pKa = 11.84SYY145 pKa = 11.06KK146 pKa = 9.63QALVGNASVKK156 pKa = 8.88EE157 pKa = 4.25TAAKK161 pKa = 9.39PAGEE165 pKa = 3.91FVAAKK170 pKa = 9.99KK171 pKa = 9.76VARR174 pKa = 11.84KK175 pKa = 8.94AATSLPQKK183 pKa = 10.42LKK185 pKa = 9.76TGNKK189 pKa = 9.25FSPLVGFARR198 pKa = 11.84AEE200 pKa = 4.11CSRR203 pKa = 11.84GFNTTTPVKK212 pKa = 9.07RR213 pKa = 11.84TYY215 pKa = 10.23PEE217 pKa = 3.95EE218 pKa = 4.02VVQTGVKK225 pKa = 9.85YY226 pKa = 9.44VSTRR230 pKa = 11.84AGHH233 pKa = 6.51QRR235 pKa = 11.84GCYY238 pKa = 8.81NAASYY243 pKa = 10.66LVEE246 pKa = 5.26FDD248 pKa = 4.4RR249 pKa = 11.84ACCNQVVEE257 pKa = 4.25QPIFEE262 pKa = 4.33KK263 pKa = 10.23RR264 pKa = 11.84TVEE267 pKa = 3.9RR268 pKa = 11.84MVVDD272 pKa = 3.65VEE274 pKa = 4.64GEE276 pKa = 4.13TPRR279 pKa = 11.84AEE281 pKa = 4.15MLKK284 pKa = 10.26SIPPRR289 pKa = 11.84ATSTTLVPVGVEE301 pKa = 3.82SLNWGNAPEE310 pKa = 4.27MAPATAKK317 pKa = 10.17RR318 pKa = 11.84PPWFGQRR325 pKa = 11.84GTTLKK330 pKa = 9.94RR331 pKa = 11.84DD332 pKa = 2.98KK333 pKa = 9.03MTRR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 4.03LSRR341 pKa = 11.84LLSALRR347 pKa = 11.84LARR350 pKa = 11.84TTVNRR355 pKa = 11.84SCSLGEE361 pKa = 3.85AFVWAWTTLGFSGAAPIPSLKK382 pKa = 9.96GWEE385 pKa = 4.11IDD387 pKa = 4.03RR388 pKa = 11.84PSPDD392 pKa = 3.38DD393 pKa = 3.23TAAPLHH399 pKa = 6.23TYY401 pKa = 8.74VV402 pKa = 4.36
MM1 pKa = 6.36TTSAVHH7 pKa = 7.01DD8 pKa = 3.74SHH10 pKa = 8.06SLRR13 pKa = 11.84KK14 pKa = 9.92GEE16 pKa = 3.85RR17 pKa = 11.84SRR19 pKa = 11.84WTRR22 pKa = 11.84RR23 pKa = 11.84TLSLSVLGPAWRR35 pKa = 11.84KK36 pKa = 8.91MGGTGALRR44 pKa = 11.84GTRR47 pKa = 11.84QRR49 pKa = 11.84GSMWVPVTVVAEE61 pKa = 4.0AVKK64 pKa = 10.43LSRR67 pKa = 11.84RR68 pKa = 11.84AHH70 pKa = 5.82YY71 pKa = 9.38SAPSCANRR79 pKa = 11.84VDD81 pKa = 3.46VRR83 pKa = 11.84AGNGRR88 pKa = 11.84KK89 pKa = 9.27IIAAVVAPATKK100 pKa = 10.0GAKK103 pKa = 9.39KK104 pKa = 10.58APLRR108 pKa = 11.84PSLVSPKK115 pKa = 9.9VVPSSRR121 pKa = 11.84GVSRR125 pKa = 11.84PSGRR129 pKa = 11.84KK130 pKa = 8.45GAYY133 pKa = 8.23SHH135 pKa = 7.38SSGTIDD141 pKa = 3.52NRR143 pKa = 11.84SYY145 pKa = 11.06KK146 pKa = 9.63QALVGNASVKK156 pKa = 8.88EE157 pKa = 4.25TAAKK161 pKa = 9.39PAGEE165 pKa = 3.91FVAAKK170 pKa = 9.99KK171 pKa = 9.76VARR174 pKa = 11.84KK175 pKa = 8.94AATSLPQKK183 pKa = 10.42LKK185 pKa = 9.76TGNKK189 pKa = 9.25FSPLVGFARR198 pKa = 11.84AEE200 pKa = 4.11CSRR203 pKa = 11.84GFNTTTPVKK212 pKa = 9.07RR213 pKa = 11.84TYY215 pKa = 10.23PEE217 pKa = 3.95EE218 pKa = 4.02VVQTGVKK225 pKa = 9.85YY226 pKa = 9.44VSTRR230 pKa = 11.84AGHH233 pKa = 6.51QRR235 pKa = 11.84GCYY238 pKa = 8.81NAASYY243 pKa = 10.66LVEE246 pKa = 5.26FDD248 pKa = 4.4RR249 pKa = 11.84ACCNQVVEE257 pKa = 4.25QPIFEE262 pKa = 4.33KK263 pKa = 10.23RR264 pKa = 11.84TVEE267 pKa = 3.9RR268 pKa = 11.84MVVDD272 pKa = 3.65VEE274 pKa = 4.64GEE276 pKa = 4.13TPRR279 pKa = 11.84AEE281 pKa = 4.15MLKK284 pKa = 10.26SIPPRR289 pKa = 11.84ATSTTLVPVGVEE301 pKa = 3.82SLNWGNAPEE310 pKa = 4.27MAPATAKK317 pKa = 10.17RR318 pKa = 11.84PPWFGQRR325 pKa = 11.84GTTLKK330 pKa = 9.94RR331 pKa = 11.84DD332 pKa = 2.98KK333 pKa = 9.03MTRR336 pKa = 11.84RR337 pKa = 11.84EE338 pKa = 4.03LSRR341 pKa = 11.84LLSALRR347 pKa = 11.84LARR350 pKa = 11.84TTVNRR355 pKa = 11.84SCSLGEE361 pKa = 3.85AFVWAWTTLGFSGAAPIPSLKK382 pKa = 9.96GWEE385 pKa = 4.11IDD387 pKa = 4.03RR388 pKa = 11.84PSPDD392 pKa = 3.38DD393 pKa = 3.23TAAPLHH399 pKa = 6.23TYY401 pKa = 8.74VV402 pKa = 4.36
Molecular weight: 43.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
402 |
402 |
402 |
402.0 |
43.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.443 ± 0.0 | 1.493 ± 0.0 |
2.239 ± 0.0 | 4.726 ± 0.0 |
2.239 ± 0.0 | 7.711 ± 0.0 |
1.493 ± 0.0 | 1.741 ± 0.0 |
6.468 ± 0.0 | 6.468 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.741 ± 0.0 | 2.736 ± 0.0 |
6.965 ± 0.0 | 1.99 ± 0.0 |
9.95 ± 0.0 | 8.955 ± 0.0 |
8.209 ± 0.0 | 9.453 ± 0.0 |
1.99 ± 0.0 | 1.99 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
