
Human papillomavirus type 48
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 2
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
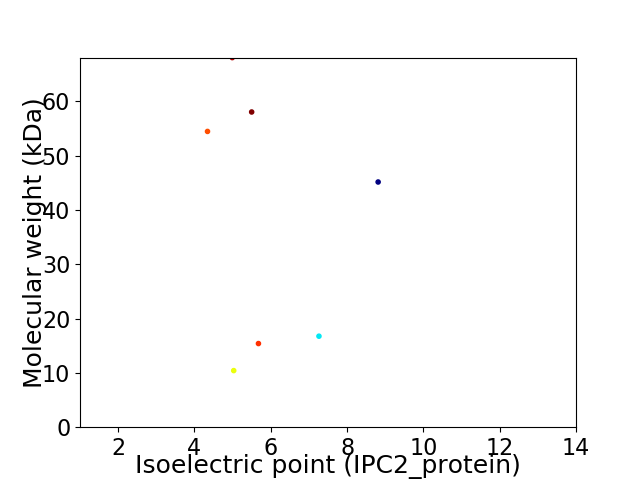
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q80925|VL2_HPV48 Minor capsid protein L2 OS=Human papillomavirus type 48 OX=40538 GN=L2 PE=3 SV=1
MM1 pKa = 7.42SLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.58RR8 pKa = 11.84ASPTDD13 pKa = 3.72LYY15 pKa = 10.02KK16 pKa = 10.6TCLQGGDD23 pKa = 4.92CIPDD27 pKa = 3.32VKK29 pKa = 11.22NKK31 pKa = 10.3FEE33 pKa = 4.39NSTIADD39 pKa = 3.73WLLKK43 pKa = 10.4IFGSLVYY50 pKa = 10.34FGNLGIGSGKK60 pKa = 10.55GSGGSFGYY68 pKa = 10.31RR69 pKa = 11.84PLGSAGSGRR78 pKa = 11.84PATDD82 pKa = 3.66LPVTRR87 pKa = 11.84PNVVIEE93 pKa = 4.36PIGPQSIVPIDD104 pKa = 4.02PGASSIVPLVEE115 pKa = 4.23GGPDD119 pKa = 2.93ISFIAPDD126 pKa = 4.17AGPGIGGEE134 pKa = 4.23DD135 pKa = 2.88IEE137 pKa = 5.67LFTFRR142 pKa = 11.84DD143 pKa = 3.46PATDD147 pKa = 3.13VGGVSGGPTTISTEE161 pKa = 3.94EE162 pKa = 4.27SEE164 pKa = 4.45TAIIDD169 pKa = 3.96ALPSATTPKK178 pKa = 10.36QLFYY182 pKa = 11.2DD183 pKa = 4.47SYY185 pKa = 10.43TQTILQTQVNPFLNNAISDD204 pKa = 4.04TNVFVDD210 pKa = 3.83PLFAGEE216 pKa = 4.44TIGDD220 pKa = 4.07NIFEE224 pKa = 4.99EE225 pKa = 5.14IPLQNLNFSFPRR237 pKa = 11.84EE238 pKa = 4.16STPVKK243 pKa = 9.65PGRR246 pKa = 11.84GLRR249 pKa = 11.84TPAQRR254 pKa = 11.84SYY256 pKa = 11.37SRR258 pKa = 11.84FMEE261 pKa = 4.14QYY263 pKa = 9.85PIQAPEE269 pKa = 4.23FLSQPSRR276 pKa = 11.84LVQFEE281 pKa = 4.31FEE283 pKa = 4.7NPAFDD288 pKa = 4.73PDD290 pKa = 3.98ISIQFQRR297 pKa = 11.84DD298 pKa = 3.7VNSLEE303 pKa = 4.24AAPNPAFADD312 pKa = 3.07IAYY315 pKa = 10.11LSRR318 pKa = 11.84PHH320 pKa = 6.36MSATSEE326 pKa = 3.96GLVRR330 pKa = 11.84VSRR333 pKa = 11.84IGSRR337 pKa = 11.84AVLQTRR343 pKa = 11.84SGLTIGPKK351 pKa = 8.11VHH353 pKa = 6.74YY354 pKa = 10.75YY355 pKa = 9.53MDD357 pKa = 4.44LSAISTEE364 pKa = 4.29AIEE367 pKa = 4.55LQTFADD373 pKa = 4.01SGHH376 pKa = 5.29VHH378 pKa = 6.81TIVDD382 pKa = 3.97DD383 pKa = 4.21FLSVTALDD391 pKa = 4.51DD392 pKa = 3.96PANIADD398 pKa = 3.61INYY401 pKa = 9.1TEE403 pKa = 6.04DD404 pKa = 4.58DD405 pKa = 4.28LLDD408 pKa = 4.27PLLEE412 pKa = 4.38NFNNSHH418 pKa = 6.0ITVQGVDD425 pKa = 3.54EE426 pKa = 4.64EE427 pKa = 5.45GEE429 pKa = 4.6TVALPIPSITNSSKK443 pKa = 9.86TFVTDD448 pKa = 2.96IAEE451 pKa = 4.39NGLFANDD458 pKa = 3.73TDD460 pKa = 4.54SLLTPASTIVPAINWFPLFDD480 pKa = 4.28SYY482 pKa = 11.97SDD484 pKa = 3.77FALDD488 pKa = 3.54PFFIPRR494 pKa = 11.84KK495 pKa = 9.42KK496 pKa = 10.16RR497 pKa = 11.84RR498 pKa = 11.84LDD500 pKa = 3.21ILL502 pKa = 3.81
MM1 pKa = 7.42SLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.58RR8 pKa = 11.84ASPTDD13 pKa = 3.72LYY15 pKa = 10.02KK16 pKa = 10.6TCLQGGDD23 pKa = 4.92CIPDD27 pKa = 3.32VKK29 pKa = 11.22NKK31 pKa = 10.3FEE33 pKa = 4.39NSTIADD39 pKa = 3.73WLLKK43 pKa = 10.4IFGSLVYY50 pKa = 10.34FGNLGIGSGKK60 pKa = 10.55GSGGSFGYY68 pKa = 10.31RR69 pKa = 11.84PLGSAGSGRR78 pKa = 11.84PATDD82 pKa = 3.66LPVTRR87 pKa = 11.84PNVVIEE93 pKa = 4.36PIGPQSIVPIDD104 pKa = 4.02PGASSIVPLVEE115 pKa = 4.23GGPDD119 pKa = 2.93ISFIAPDD126 pKa = 4.17AGPGIGGEE134 pKa = 4.23DD135 pKa = 2.88IEE137 pKa = 5.67LFTFRR142 pKa = 11.84DD143 pKa = 3.46PATDD147 pKa = 3.13VGGVSGGPTTISTEE161 pKa = 3.94EE162 pKa = 4.27SEE164 pKa = 4.45TAIIDD169 pKa = 3.96ALPSATTPKK178 pKa = 10.36QLFYY182 pKa = 11.2DD183 pKa = 4.47SYY185 pKa = 10.43TQTILQTQVNPFLNNAISDD204 pKa = 4.04TNVFVDD210 pKa = 3.83PLFAGEE216 pKa = 4.44TIGDD220 pKa = 4.07NIFEE224 pKa = 4.99EE225 pKa = 5.14IPLQNLNFSFPRR237 pKa = 11.84EE238 pKa = 4.16STPVKK243 pKa = 9.65PGRR246 pKa = 11.84GLRR249 pKa = 11.84TPAQRR254 pKa = 11.84SYY256 pKa = 11.37SRR258 pKa = 11.84FMEE261 pKa = 4.14QYY263 pKa = 9.85PIQAPEE269 pKa = 4.23FLSQPSRR276 pKa = 11.84LVQFEE281 pKa = 4.31FEE283 pKa = 4.7NPAFDD288 pKa = 4.73PDD290 pKa = 3.98ISIQFQRR297 pKa = 11.84DD298 pKa = 3.7VNSLEE303 pKa = 4.24AAPNPAFADD312 pKa = 3.07IAYY315 pKa = 10.11LSRR318 pKa = 11.84PHH320 pKa = 6.36MSATSEE326 pKa = 3.96GLVRR330 pKa = 11.84VSRR333 pKa = 11.84IGSRR337 pKa = 11.84AVLQTRR343 pKa = 11.84SGLTIGPKK351 pKa = 8.11VHH353 pKa = 6.74YY354 pKa = 10.75YY355 pKa = 9.53MDD357 pKa = 4.44LSAISTEE364 pKa = 4.29AIEE367 pKa = 4.55LQTFADD373 pKa = 4.01SGHH376 pKa = 5.29VHH378 pKa = 6.81TIVDD382 pKa = 3.97DD383 pKa = 4.21FLSVTALDD391 pKa = 4.51DD392 pKa = 3.96PANIADD398 pKa = 3.61INYY401 pKa = 9.1TEE403 pKa = 6.04DD404 pKa = 4.58DD405 pKa = 4.28LLDD408 pKa = 4.27PLLEE412 pKa = 4.38NFNNSHH418 pKa = 6.0ITVQGVDD425 pKa = 3.54EE426 pKa = 4.64EE427 pKa = 5.45GEE429 pKa = 4.6TVALPIPSITNSSKK443 pKa = 9.86TFVTDD448 pKa = 2.96IAEE451 pKa = 4.39NGLFANDD458 pKa = 3.73TDD460 pKa = 4.54SLLTPASTIVPAINWFPLFDD480 pKa = 4.28SYY482 pKa = 11.97SDD484 pKa = 3.77FALDD488 pKa = 3.54PFFIPRR494 pKa = 11.84KK495 pKa = 9.42KK496 pKa = 10.16RR497 pKa = 11.84RR498 pKa = 11.84LDD500 pKa = 3.21ILL502 pKa = 3.81
Molecular weight: 54.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q80924|Q80924_HPV48 E4 protein (Fragment) OS=Human papillomavirus type 48 OX=40538 GN=E4 PE=4 SV=1
MM1 pKa = 7.3QPEE4 pKa = 4.43TQEE7 pKa = 4.11SLSTRR12 pKa = 11.84FAAQQEE18 pKa = 4.39IQLTLIEE25 pKa = 4.6KK26 pKa = 10.21EE27 pKa = 4.18SYY29 pKa = 10.22DD30 pKa = 4.36LKK32 pKa = 11.42DD33 pKa = 3.52HH34 pKa = 6.49LAYY37 pKa = 9.5WKK39 pKa = 10.38AVRR42 pKa = 11.84LEE44 pKa = 3.95NVIAYY49 pKa = 7.14YY50 pKa = 10.47ARR52 pKa = 11.84KK53 pKa = 8.84EE54 pKa = 4.2HH55 pKa = 5.73ITKK58 pKa = 10.61LGLQPLPTLAVTEE71 pKa = 4.46YY72 pKa = 10.25KK73 pKa = 10.63AKK75 pKa = 10.27EE76 pKa = 4.14AINIQLLIQSLLKK89 pKa = 10.69SEE91 pKa = 4.67FALEE95 pKa = 3.83RR96 pKa = 11.84WTLAEE101 pKa = 4.27TSAEE105 pKa = 4.39TINSSPRR112 pKa = 11.84NCFKK116 pKa = 10.74KK117 pKa = 10.61VPFIVNVWFDD127 pKa = 3.29NDD129 pKa = 3.34EE130 pKa = 4.48RR131 pKa = 11.84NSFPYY136 pKa = 9.63TCWDD140 pKa = 4.43FIYY143 pKa = 11.11YY144 pKa = 9.93QDD146 pKa = 5.56DD147 pKa = 3.4QNKK150 pKa = 5.75WHH152 pKa = 6.26KK153 pKa = 9.71TEE155 pKa = 4.47GLVDD159 pKa = 3.92HH160 pKa = 6.8NGCYY164 pKa = 10.01YY165 pKa = 11.06VDD167 pKa = 4.08LNGDD171 pKa = 3.98FVYY174 pKa = 8.72FTLFQPDD181 pKa = 3.17AVKK184 pKa = 10.75YY185 pKa = 10.21GKK187 pKa = 8.56TGLWTVRR194 pKa = 11.84FKK196 pKa = 11.58NKK198 pKa = 9.28TISASVTSSSRR209 pKa = 11.84NTNPSSEE216 pKa = 4.21SRR218 pKa = 11.84VGLSTSSSSEE228 pKa = 3.86SPRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84PSISEE238 pKa = 3.8NSNTEE243 pKa = 3.99SPTSSTSRR251 pKa = 11.84LRR253 pKa = 11.84EE254 pKa = 3.48RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84EE259 pKa = 3.56PRR261 pKa = 11.84EE262 pKa = 3.86SGTTDD267 pKa = 2.85TTPRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GTKK276 pKa = 9.56RR277 pKa = 11.84KK278 pKa = 9.53LGSDD282 pKa = 3.64SAPTPSEE289 pKa = 3.86VGSRR293 pKa = 11.84STTLARR299 pKa = 11.84HH300 pKa = 6.24GYY302 pKa = 9.26SRR304 pKa = 11.84LGRR307 pKa = 11.84LQEE310 pKa = 4.06EE311 pKa = 4.44ARR313 pKa = 11.84DD314 pKa = 3.81PPLVLFTGQQNNLKK328 pKa = 9.65CWRR331 pKa = 11.84NRR333 pKa = 11.84CTTKK337 pKa = 10.56YY338 pKa = 10.96ASLFLCFSSVWKK350 pKa = 9.68WLGPNSDD357 pKa = 3.27GGAAKK362 pKa = 10.53VLVAFKK368 pKa = 10.83SDD370 pKa = 2.94AQRR373 pKa = 11.84QVFLNTVHH381 pKa = 6.59IPKK384 pKa = 9.03GTTITLGRR392 pKa = 11.84LDD394 pKa = 3.68SLL396 pKa = 4.32
MM1 pKa = 7.3QPEE4 pKa = 4.43TQEE7 pKa = 4.11SLSTRR12 pKa = 11.84FAAQQEE18 pKa = 4.39IQLTLIEE25 pKa = 4.6KK26 pKa = 10.21EE27 pKa = 4.18SYY29 pKa = 10.22DD30 pKa = 4.36LKK32 pKa = 11.42DD33 pKa = 3.52HH34 pKa = 6.49LAYY37 pKa = 9.5WKK39 pKa = 10.38AVRR42 pKa = 11.84LEE44 pKa = 3.95NVIAYY49 pKa = 7.14YY50 pKa = 10.47ARR52 pKa = 11.84KK53 pKa = 8.84EE54 pKa = 4.2HH55 pKa = 5.73ITKK58 pKa = 10.61LGLQPLPTLAVTEE71 pKa = 4.46YY72 pKa = 10.25KK73 pKa = 10.63AKK75 pKa = 10.27EE76 pKa = 4.14AINIQLLIQSLLKK89 pKa = 10.69SEE91 pKa = 4.67FALEE95 pKa = 3.83RR96 pKa = 11.84WTLAEE101 pKa = 4.27TSAEE105 pKa = 4.39TINSSPRR112 pKa = 11.84NCFKK116 pKa = 10.74KK117 pKa = 10.61VPFIVNVWFDD127 pKa = 3.29NDD129 pKa = 3.34EE130 pKa = 4.48RR131 pKa = 11.84NSFPYY136 pKa = 9.63TCWDD140 pKa = 4.43FIYY143 pKa = 11.11YY144 pKa = 9.93QDD146 pKa = 5.56DD147 pKa = 3.4QNKK150 pKa = 5.75WHH152 pKa = 6.26KK153 pKa = 9.71TEE155 pKa = 4.47GLVDD159 pKa = 3.92HH160 pKa = 6.8NGCYY164 pKa = 10.01YY165 pKa = 11.06VDD167 pKa = 4.08LNGDD171 pKa = 3.98FVYY174 pKa = 8.72FTLFQPDD181 pKa = 3.17AVKK184 pKa = 10.75YY185 pKa = 10.21GKK187 pKa = 8.56TGLWTVRR194 pKa = 11.84FKK196 pKa = 11.58NKK198 pKa = 9.28TISASVTSSSRR209 pKa = 11.84NTNPSSEE216 pKa = 4.21SRR218 pKa = 11.84VGLSTSSSSEE228 pKa = 3.86SPRR231 pKa = 11.84RR232 pKa = 11.84RR233 pKa = 11.84PSISEE238 pKa = 3.8NSNTEE243 pKa = 3.99SPTSSTSRR251 pKa = 11.84LRR253 pKa = 11.84EE254 pKa = 3.48RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84EE259 pKa = 3.56PRR261 pKa = 11.84EE262 pKa = 3.86SGTTDD267 pKa = 2.85TTPRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84GTKK276 pKa = 9.56RR277 pKa = 11.84KK278 pKa = 9.53LGSDD282 pKa = 3.64SAPTPSEE289 pKa = 3.86VGSRR293 pKa = 11.84STTLARR299 pKa = 11.84HH300 pKa = 6.24GYY302 pKa = 9.26SRR304 pKa = 11.84LGRR307 pKa = 11.84LQEE310 pKa = 4.06EE311 pKa = 4.44ARR313 pKa = 11.84DD314 pKa = 3.81PPLVLFTGQQNNLKK328 pKa = 9.65CWRR331 pKa = 11.84NRR333 pKa = 11.84CTTKK337 pKa = 10.56YY338 pKa = 10.96ASLFLCFSSVWKK350 pKa = 9.68WLGPNSDD357 pKa = 3.27GGAAKK362 pKa = 10.53VLVAFKK368 pKa = 10.83SDD370 pKa = 2.94AQRR373 pKa = 11.84QVFLNTVHH381 pKa = 6.59IPKK384 pKa = 9.03GTTITLGRR392 pKa = 11.84LDD394 pKa = 3.68SLL396 pKa = 4.32
Molecular weight: 45.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2372 |
93 |
593 |
338.9 |
38.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.818 ± 0.338 | 2.445 ± 0.752 |
6.703 ± 0.487 | 6.282 ± 0.497 |
5.143 ± 0.559 | 5.228 ± 0.84 |
1.56 ± 0.225 | 5.228 ± 0.736 |
5.396 ± 0.825 | 9.612 ± 0.482 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.391 ± 0.412 | 5.228 ± 0.33 |
5.776 ± 0.922 | 3.71 ± 0.264 |
5.649 ± 0.888 | 7.715 ± 0.697 |
6.998 ± 0.409 | 5.438 ± 0.416 |
1.391 ± 0.271 | 3.288 ± 0.511 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
