
Grapevine enamovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Enamovirus
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
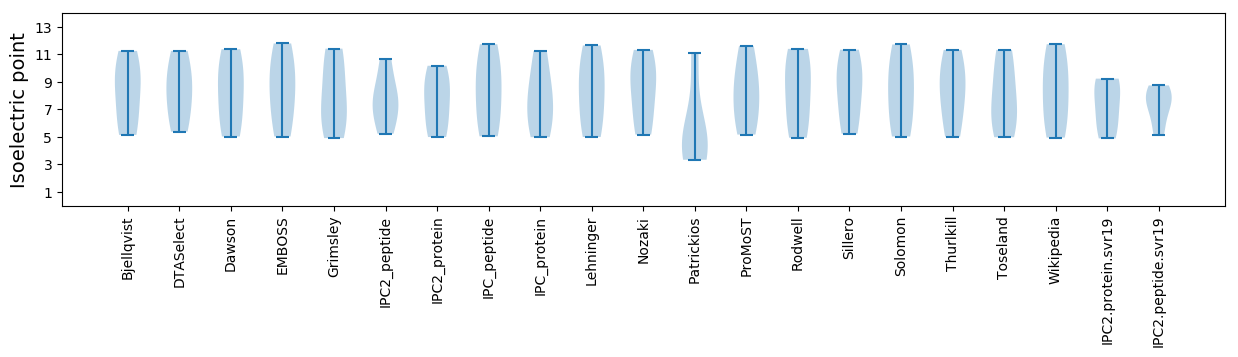
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9YLK7|A0A1X9YLK7_9LUTE Serine protease OS=Grapevine enamovirus 1 OX=2560515 PE=4 SV=1
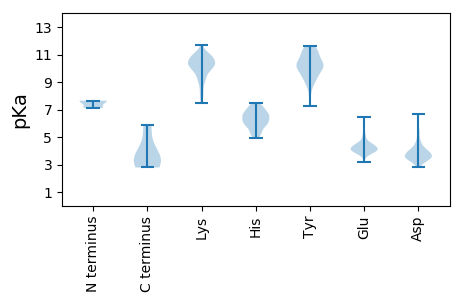
MM1 pKa = 7.58SSEE4 pKa = 4.15GNSEE8 pKa = 3.65IKK10 pKa = 10.33KK11 pKa = 9.66ICEE14 pKa = 3.67QSKK17 pKa = 9.23GFSFSRR23 pKa = 11.84CDD25 pKa = 3.54LNSRR29 pKa = 11.84DD30 pKa = 4.74FLIEE34 pKa = 3.88SCEE37 pKa = 4.08KK38 pKa = 9.73ILAVLEE44 pKa = 4.34HH45 pKa = 6.78HH46 pKa = 7.17VSQPDD51 pKa = 3.45SHH53 pKa = 7.46HH54 pKa = 7.24DD55 pKa = 3.5SCFTAFLYY63 pKa = 7.7FACLLMLTHH72 pKa = 6.95PASWCSASRR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 5.91LAYY86 pKa = 10.37DD87 pKa = 3.3ATLLGGFPLICGFRR101 pKa = 11.84AEE103 pKa = 5.58DD104 pKa = 3.76YY105 pKa = 11.42DD106 pKa = 4.3FMGRR110 pKa = 11.84LGARR114 pKa = 11.84FGVINSEE121 pKa = 4.34PIAVHH126 pKa = 5.33TAQGRR131 pKa = 11.84GAGIPYY137 pKa = 10.34AGLCNFRR144 pKa = 11.84NEE146 pKa = 4.12SEE148 pKa = 4.28RR149 pKa = 11.84LFAYY153 pKa = 9.49VGWYY157 pKa = 9.03LHH159 pKa = 7.4LYY161 pKa = 7.22RR162 pKa = 11.84TSNGGNSVQVVTRR175 pKa = 11.84LQNNAVSHH183 pKa = 5.03VLSRR187 pKa = 11.84INPRR191 pKa = 11.84DD192 pKa = 3.22PADD195 pKa = 3.89FMWGFARR202 pKa = 11.84VATMANRR209 pKa = 11.84RR210 pKa = 11.84PPHH213 pKa = 5.39MLRR216 pKa = 11.84SRR218 pKa = 11.84ADD220 pKa = 3.09NVLLVRR226 pKa = 11.84RR227 pKa = 11.84LFDD230 pKa = 2.93ICRR233 pKa = 11.84GDD235 pKa = 3.74VQTDD239 pKa = 3.24DD240 pKa = 4.21HH241 pKa = 7.19HH242 pKa = 7.22DD243 pKa = 3.69LNGYY247 pKa = 8.99GLASLLGNITTDD259 pKa = 3.71CAVSCPAPYY268 pKa = 10.41VADD271 pKa = 3.99LDD273 pKa = 3.97YY274 pKa = 10.84PYY276 pKa = 10.41IDD278 pKa = 3.75TLGLTDD284 pKa = 4.85SDD286 pKa = 4.47EE287 pKa = 6.48DD288 pKa = 4.42DD289 pKa = 5.23DD290 pKa = 6.68LDD292 pKa = 3.92INEE295 pKa = 5.24AGVEE299 pKa = 4.25VPPEE303 pKa = 3.94IEE305 pKa = 4.05PDD307 pKa = 2.86NWGGLFPP314 pKa = 5.91
MM1 pKa = 7.58SSEE4 pKa = 4.15GNSEE8 pKa = 3.65IKK10 pKa = 10.33KK11 pKa = 9.66ICEE14 pKa = 3.67QSKK17 pKa = 9.23GFSFSRR23 pKa = 11.84CDD25 pKa = 3.54LNSRR29 pKa = 11.84DD30 pKa = 4.74FLIEE34 pKa = 3.88SCEE37 pKa = 4.08KK38 pKa = 9.73ILAVLEE44 pKa = 4.34HH45 pKa = 6.78HH46 pKa = 7.17VSQPDD51 pKa = 3.45SHH53 pKa = 7.46HH54 pKa = 7.24DD55 pKa = 3.5SCFTAFLYY63 pKa = 7.7FACLLMLTHH72 pKa = 6.95PASWCSASRR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 5.91LAYY86 pKa = 10.37DD87 pKa = 3.3ATLLGGFPLICGFRR101 pKa = 11.84AEE103 pKa = 5.58DD104 pKa = 3.76YY105 pKa = 11.42DD106 pKa = 4.3FMGRR110 pKa = 11.84LGARR114 pKa = 11.84FGVINSEE121 pKa = 4.34PIAVHH126 pKa = 5.33TAQGRR131 pKa = 11.84GAGIPYY137 pKa = 10.34AGLCNFRR144 pKa = 11.84NEE146 pKa = 4.12SEE148 pKa = 4.28RR149 pKa = 11.84LFAYY153 pKa = 9.49VGWYY157 pKa = 9.03LHH159 pKa = 7.4LYY161 pKa = 7.22RR162 pKa = 11.84TSNGGNSVQVVTRR175 pKa = 11.84LQNNAVSHH183 pKa = 5.03VLSRR187 pKa = 11.84INPRR191 pKa = 11.84DD192 pKa = 3.22PADD195 pKa = 3.89FMWGFARR202 pKa = 11.84VATMANRR209 pKa = 11.84RR210 pKa = 11.84PPHH213 pKa = 5.39MLRR216 pKa = 11.84SRR218 pKa = 11.84ADD220 pKa = 3.09NVLLVRR226 pKa = 11.84RR227 pKa = 11.84LFDD230 pKa = 2.93ICRR233 pKa = 11.84GDD235 pKa = 3.74VQTDD239 pKa = 3.24DD240 pKa = 4.21HH241 pKa = 7.19HH242 pKa = 7.22DD243 pKa = 3.69LNGYY247 pKa = 8.99GLASLLGNITTDD259 pKa = 3.71CAVSCPAPYY268 pKa = 10.41VADD271 pKa = 3.99LDD273 pKa = 3.97YY274 pKa = 10.84PYY276 pKa = 10.41IDD278 pKa = 3.75TLGLTDD284 pKa = 4.85SDD286 pKa = 4.47EE287 pKa = 6.48DD288 pKa = 4.42DD289 pKa = 5.23DD290 pKa = 6.68LDD292 pKa = 3.92INEE295 pKa = 5.24AGVEE299 pKa = 4.25VPPEE303 pKa = 3.94IEE305 pKa = 4.05PDD307 pKa = 2.86NWGGLFPP314 pKa = 5.91
Molecular weight: 34.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9YLK6|A0A1X9YLK6_9LUTE p0 protein OS=Grapevine enamovirus 1 OX=2560515 PE=4 SV=1
MM1 pKa = 7.11VARR4 pKa = 11.84SKK6 pKa = 10.98KK7 pKa = 9.87AGKK10 pKa = 9.62KK11 pKa = 9.8KK12 pKa = 10.57SAGPGNRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84LQPRR25 pKa = 11.84ARR27 pKa = 11.84QMVVVSTQRR36 pKa = 11.84PRR38 pKa = 11.84KK39 pKa = 8.01PRR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84TRR46 pKa = 11.84GRR48 pKa = 11.84GSQGGGLNAGVNFTFLVNSFAGNASGTIKK77 pKa = 10.65FGPNLTEE84 pKa = 3.62SSAFVGVLGSFQRR97 pKa = 11.84YY98 pKa = 9.25RR99 pKa = 11.84IVSLQVHH106 pKa = 5.89YY107 pKa = 10.09VTEE110 pKa = 4.22ASKK113 pKa = 10.31MDD115 pKa = 3.94RR116 pKa = 11.84GCIAYY121 pKa = 9.45HH122 pKa = 6.36VDD124 pKa = 3.73TSCSMRR130 pKa = 11.84ASGLLPTTSWPVTQSALKK148 pKa = 9.28TYY150 pKa = 10.12GAGVLGDD157 pKa = 3.66QPHH160 pKa = 6.09YY161 pKa = 10.77EE162 pKa = 4.06HH163 pKa = 6.79TKK165 pKa = 7.7EE166 pKa = 4.46QFWFLYY172 pKa = 9.7KK173 pKa = 11.0GNGSSDD179 pKa = 3.07IAGHH183 pKa = 6.18LRR185 pKa = 11.84LTIRR189 pKa = 11.84VVFTNTLL196 pKa = 3.23
MM1 pKa = 7.11VARR4 pKa = 11.84SKK6 pKa = 10.98KK7 pKa = 9.87AGKK10 pKa = 9.62KK11 pKa = 9.8KK12 pKa = 10.57SAGPGNRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84LQPRR25 pKa = 11.84ARR27 pKa = 11.84QMVVVSTQRR36 pKa = 11.84PRR38 pKa = 11.84KK39 pKa = 8.01PRR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84TRR46 pKa = 11.84GRR48 pKa = 11.84GSQGGGLNAGVNFTFLVNSFAGNASGTIKK77 pKa = 10.65FGPNLTEE84 pKa = 3.62SSAFVGVLGSFQRR97 pKa = 11.84YY98 pKa = 9.25RR99 pKa = 11.84IVSLQVHH106 pKa = 5.89YY107 pKa = 10.09VTEE110 pKa = 4.22ASKK113 pKa = 10.31MDD115 pKa = 3.94RR116 pKa = 11.84GCIAYY121 pKa = 9.45HH122 pKa = 6.36VDD124 pKa = 3.73TSCSMRR130 pKa = 11.84ASGLLPTTSWPVTQSALKK148 pKa = 9.28TYY150 pKa = 10.12GAGVLGDD157 pKa = 3.66QPHH160 pKa = 6.09YY161 pKa = 10.77EE162 pKa = 4.06HH163 pKa = 6.79TKK165 pKa = 7.7EE166 pKa = 4.46QFWFLYY172 pKa = 9.7KK173 pKa = 11.0GNGSSDD179 pKa = 3.07IAGHH183 pKa = 6.18LRR185 pKa = 11.84LTIRR189 pKa = 11.84VVFTNTLL196 pKa = 3.23
Molecular weight: 21.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3098 |
196 |
1231 |
619.6 |
68.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.23 ± 0.334 | 2.324 ± 0.251 |
5.003 ± 0.425 | 5.1 ± 0.432 |
3.777 ± 0.272 | 8.102 ± 0.416 |
2.227 ± 0.251 | 4.229 ± 0.266 |
5.229 ± 0.755 | 8.489 ± 0.838 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.744 ± 0.258 | 3.163 ± 0.359 |
6.359 ± 0.835 | 4.099 ± 0.319 |
5.939 ± 0.661 | 8.522 ± 0.18 |
5.875 ± 0.486 | 6.746 ± 0.215 |
2.195 ± 0.358 | 2.615 ± 0.171 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
