
Rotavirus G chicken/03V0567/DEU/2003
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus G
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
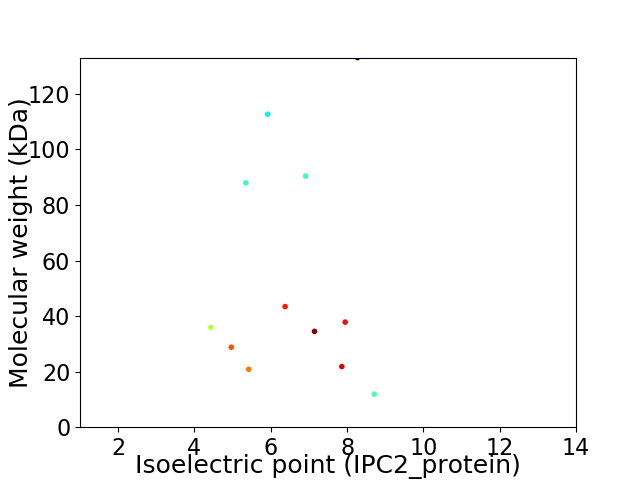
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4H298|M4H298_9REOV NSP1-1 OS=Rotavirus G chicken/03V0567/DEU/2003 OX=994995 PE=4 SV=1
MM1 pKa = 7.92AEE3 pKa = 4.04LLKK6 pKa = 11.11VCFDD10 pKa = 3.87EE11 pKa = 4.62MMKK14 pKa = 10.45SGNCSFEE21 pKa = 3.95HH22 pKa = 7.57LMEE25 pKa = 4.63KK26 pKa = 10.21MNDD29 pKa = 3.06AGIDD33 pKa = 3.54LDD35 pKa = 3.43KK36 pKa = 10.66WKK38 pKa = 10.45QAFDD42 pKa = 3.8GARR45 pKa = 11.84LPQRR49 pKa = 11.84MTKK52 pKa = 8.59STLAIQNTNLEE63 pKa = 4.29KK64 pKa = 10.61EE65 pKa = 4.32IVMLRR70 pKa = 11.84AKK72 pKa = 7.54EE73 pKa = 3.99HH74 pKa = 5.41RR75 pKa = 11.84HH76 pKa = 6.13GYY78 pKa = 10.03DD79 pKa = 2.79KK80 pKa = 11.36NKK82 pKa = 9.0RR83 pKa = 11.84TLANFDD89 pKa = 3.67VEE91 pKa = 4.44RR92 pKa = 11.84KK93 pKa = 9.48NGNTILTPKK102 pKa = 9.19TRR104 pKa = 11.84LAEE107 pKa = 4.73IILLNSTRR115 pKa = 11.84DD116 pKa = 3.8LKK118 pKa = 11.32LSAPPSDD125 pKa = 4.06YY126 pKa = 10.76IEE128 pKa = 4.2EE129 pKa = 4.84LEE131 pKa = 4.72DD132 pKa = 4.14KK133 pKa = 10.61VAQLEE138 pKa = 4.21EE139 pKa = 4.05EE140 pKa = 4.35RR141 pKa = 11.84LNLMRR146 pKa = 11.84CNNDD150 pKa = 2.0IYY152 pKa = 10.89YY153 pKa = 10.44QYY155 pKa = 10.34LTLHH159 pKa = 5.1TTSYY163 pKa = 11.15CLQEE167 pKa = 3.96TNKK170 pKa = 8.98WQEE173 pKa = 3.78KK174 pKa = 9.03MISRR178 pKa = 11.84LQEE181 pKa = 3.64NEE183 pKa = 3.63MVMSSEE189 pKa = 3.92INDD192 pKa = 4.01LKK194 pKa = 11.68SMVRR198 pKa = 11.84TLTRR202 pKa = 11.84EE203 pKa = 3.81LNYY206 pKa = 10.35EE207 pKa = 3.68ISFEE211 pKa = 4.3DD212 pKa = 4.37EE213 pKa = 4.16PDD215 pKa = 2.85TGYY218 pKa = 10.56ISDD221 pKa = 4.21EE222 pKa = 3.97NSQVSEE228 pKa = 4.65DD229 pKa = 4.08DD230 pKa = 4.27LSEE233 pKa = 4.25QEE235 pKa = 5.27LDD237 pKa = 4.55NDD239 pKa = 3.68QQDD242 pKa = 3.96EE243 pKa = 4.5NEE245 pKa = 4.42SSDD248 pKa = 3.66EE249 pKa = 4.41EE250 pKa = 4.25IFEE253 pKa = 5.65DD254 pKa = 5.78LDD256 pKa = 4.21DD257 pKa = 6.53LINQYY262 pKa = 7.07EE263 pKa = 4.01QRR265 pKa = 11.84QRR267 pKa = 11.84LMEE270 pKa = 4.03RR271 pKa = 11.84EE272 pKa = 4.11RR273 pKa = 11.84ILNEE277 pKa = 3.89LQAPDD282 pKa = 4.38YY283 pKa = 10.36DD284 pKa = 4.28TEE286 pKa = 4.29TDD288 pKa = 3.96DD289 pKa = 4.12EE290 pKa = 4.92NEE292 pKa = 3.73YY293 pKa = 10.94AYY295 pKa = 10.84NIEE298 pKa = 4.1RR299 pKa = 11.84EE300 pKa = 4.2HH301 pKa = 6.53RR302 pKa = 11.84HH303 pKa = 4.81
MM1 pKa = 7.92AEE3 pKa = 4.04LLKK6 pKa = 11.11VCFDD10 pKa = 3.87EE11 pKa = 4.62MMKK14 pKa = 10.45SGNCSFEE21 pKa = 3.95HH22 pKa = 7.57LMEE25 pKa = 4.63KK26 pKa = 10.21MNDD29 pKa = 3.06AGIDD33 pKa = 3.54LDD35 pKa = 3.43KK36 pKa = 10.66WKK38 pKa = 10.45QAFDD42 pKa = 3.8GARR45 pKa = 11.84LPQRR49 pKa = 11.84MTKK52 pKa = 8.59STLAIQNTNLEE63 pKa = 4.29KK64 pKa = 10.61EE65 pKa = 4.32IVMLRR70 pKa = 11.84AKK72 pKa = 7.54EE73 pKa = 3.99HH74 pKa = 5.41RR75 pKa = 11.84HH76 pKa = 6.13GYY78 pKa = 10.03DD79 pKa = 2.79KK80 pKa = 11.36NKK82 pKa = 9.0RR83 pKa = 11.84TLANFDD89 pKa = 3.67VEE91 pKa = 4.44RR92 pKa = 11.84KK93 pKa = 9.48NGNTILTPKK102 pKa = 9.19TRR104 pKa = 11.84LAEE107 pKa = 4.73IILLNSTRR115 pKa = 11.84DD116 pKa = 3.8LKK118 pKa = 11.32LSAPPSDD125 pKa = 4.06YY126 pKa = 10.76IEE128 pKa = 4.2EE129 pKa = 4.84LEE131 pKa = 4.72DD132 pKa = 4.14KK133 pKa = 10.61VAQLEE138 pKa = 4.21EE139 pKa = 4.05EE140 pKa = 4.35RR141 pKa = 11.84LNLMRR146 pKa = 11.84CNNDD150 pKa = 2.0IYY152 pKa = 10.89YY153 pKa = 10.44QYY155 pKa = 10.34LTLHH159 pKa = 5.1TTSYY163 pKa = 11.15CLQEE167 pKa = 3.96TNKK170 pKa = 8.98WQEE173 pKa = 3.78KK174 pKa = 9.03MISRR178 pKa = 11.84LQEE181 pKa = 3.64NEE183 pKa = 3.63MVMSSEE189 pKa = 3.92INDD192 pKa = 4.01LKK194 pKa = 11.68SMVRR198 pKa = 11.84TLTRR202 pKa = 11.84EE203 pKa = 3.81LNYY206 pKa = 10.35EE207 pKa = 3.68ISFEE211 pKa = 4.3DD212 pKa = 4.37EE213 pKa = 4.16PDD215 pKa = 2.85TGYY218 pKa = 10.56ISDD221 pKa = 4.21EE222 pKa = 3.97NSQVSEE228 pKa = 4.65DD229 pKa = 4.08DD230 pKa = 4.27LSEE233 pKa = 4.25QEE235 pKa = 5.27LDD237 pKa = 4.55NDD239 pKa = 3.68QQDD242 pKa = 3.96EE243 pKa = 4.5NEE245 pKa = 4.42SSDD248 pKa = 3.66EE249 pKa = 4.41EE250 pKa = 4.25IFEE253 pKa = 5.65DD254 pKa = 5.78LDD256 pKa = 4.21DD257 pKa = 6.53LINQYY262 pKa = 7.07EE263 pKa = 4.01QRR265 pKa = 11.84QRR267 pKa = 11.84LMEE270 pKa = 4.03RR271 pKa = 11.84EE272 pKa = 4.11RR273 pKa = 11.84ILNEE277 pKa = 3.89LQAPDD282 pKa = 4.38YY283 pKa = 10.36DD284 pKa = 4.28TEE286 pKa = 4.29TDD288 pKa = 3.96DD289 pKa = 4.12EE290 pKa = 4.92NEE292 pKa = 3.73YY293 pKa = 10.94AYY295 pKa = 10.84NIEE298 pKa = 4.1RR299 pKa = 11.84EE300 pKa = 4.2HH301 pKa = 6.53RR302 pKa = 11.84HH303 pKa = 4.81
Molecular weight: 35.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4H2A1|M4H2A1_9REOV NSP5 OS=Rotavirus G chicken/03V0567/DEU/2003 OX=994995 PE=4 SV=1
MM1 pKa = 7.5GNSNTNIQVSQQNTHH16 pKa = 5.4IQASDD21 pKa = 3.43SKK23 pKa = 11.61LEE25 pKa = 3.95LHH27 pKa = 6.46DD28 pKa = 4.03QKK30 pKa = 10.04TATLQSTQLLISIGAIIIVALILLLLFSLILNCYY64 pKa = 9.05LCNRR68 pKa = 11.84LKK70 pKa = 10.83KK71 pKa = 10.49KK72 pKa = 10.44NGYY75 pKa = 9.13LKK77 pKa = 10.56RR78 pKa = 11.84EE79 pKa = 3.96RR80 pKa = 11.84KK81 pKa = 9.26ISNLRR86 pKa = 11.84DD87 pKa = 3.05KK88 pKa = 11.47GLDD91 pKa = 3.24KK92 pKa = 11.13FILDD96 pKa = 4.27KK97 pKa = 11.37PNDD100 pKa = 3.62ITSGCVV106 pKa = 2.7
MM1 pKa = 7.5GNSNTNIQVSQQNTHH16 pKa = 5.4IQASDD21 pKa = 3.43SKK23 pKa = 11.61LEE25 pKa = 3.95LHH27 pKa = 6.46DD28 pKa = 4.03QKK30 pKa = 10.04TATLQSTQLLISIGAIIIVALILLLLFSLILNCYY64 pKa = 9.05LCNRR68 pKa = 11.84LKK70 pKa = 10.83KK71 pKa = 10.49KK72 pKa = 10.44NGYY75 pKa = 9.13LKK77 pKa = 10.56RR78 pKa = 11.84EE79 pKa = 3.96RR80 pKa = 11.84KK81 pKa = 9.26ISNLRR86 pKa = 11.84DD87 pKa = 3.05KK88 pKa = 11.47GLDD91 pKa = 3.24KK92 pKa = 11.13FILDD96 pKa = 4.27KK97 pKa = 11.37PNDD100 pKa = 3.62ITSGCVV106 pKa = 2.7
Molecular weight: 11.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5730 |
106 |
1160 |
477.5 |
54.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.637 ± 0.503 | 1.134 ± 0.304 |
6.754 ± 0.38 | 6.859 ± 0.674 |
3.787 ± 0.369 | 4.188 ± 0.33 |
1.518 ± 0.199 | 8.133 ± 0.319 |
7.818 ± 0.455 | 7.784 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.264 ± 0.283 | 6.3 ± 0.407 |
3.246 ± 0.281 | 4.206 ± 0.297 |
5.34 ± 0.341 | 6.405 ± 0.263 |
6.073 ± 0.315 | 6.318 ± 0.468 |
0.96 ± 0.162 | 4.276 ± 0.532 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
