
Penicillium chrysogenum virus (isolate Caston/2003) (PcV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Alphachrysovirus; Penicillium chrysogenum virus
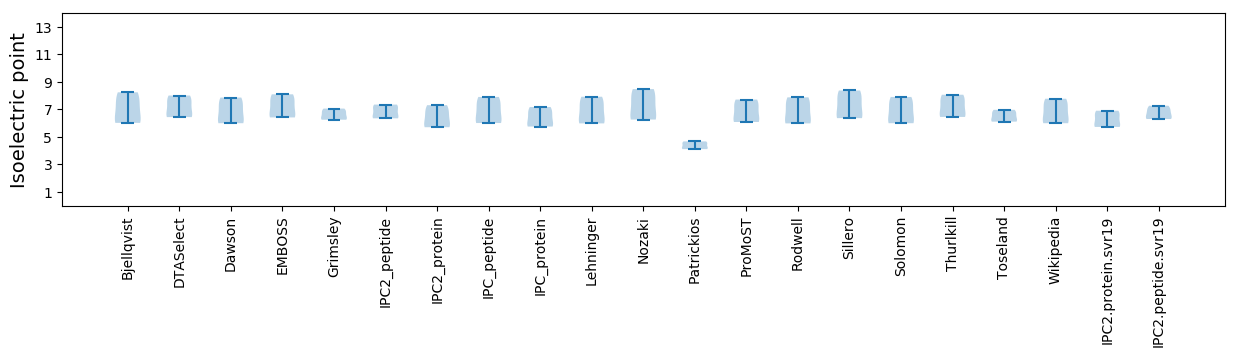
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
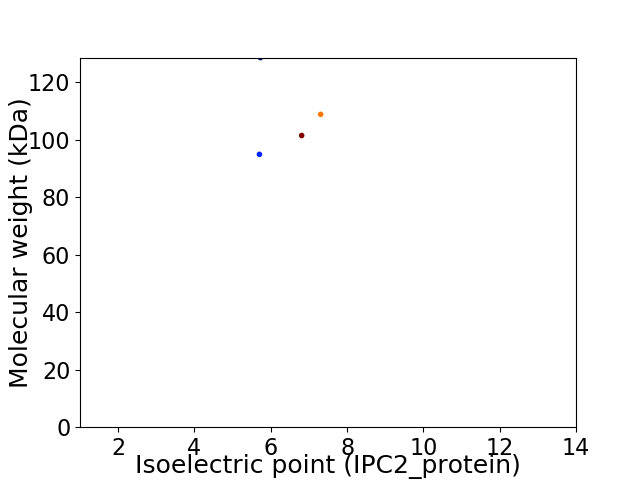
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8JVC0|P3_PCVC Uncharacterized protein p3 OS=Penicillium chrysogenum virus (isolate Caston/2003) OX=654932 GN=p3 PE=4 SV=1
MM1 pKa = 7.24AAEE4 pKa = 4.4ARR6 pKa = 11.84VIDD9 pKa = 4.24YY10 pKa = 8.19MKK12 pKa = 10.71GLPQSYY18 pKa = 6.77QHH20 pKa = 6.8RR21 pKa = 11.84EE22 pKa = 3.69TYY24 pKa = 10.4FEE26 pKa = 4.74QFLEE30 pKa = 4.34QNSEE34 pKa = 4.16RR35 pKa = 11.84LVEE38 pKa = 4.75EE39 pKa = 3.44IDD41 pKa = 3.91GPVVEE46 pKa = 5.52KK47 pKa = 11.11GLISDD52 pKa = 4.37GEE54 pKa = 4.23KK55 pKa = 10.56EE56 pKa = 4.42IIRR59 pKa = 11.84HH60 pKa = 5.96LNVQDD65 pKa = 3.46KK66 pKa = 8.36FTINGFRR73 pKa = 11.84KK74 pKa = 9.47SVKK77 pKa = 8.0ICHH80 pKa = 5.5MVSLGATRR88 pKa = 11.84QTKK91 pKa = 8.55RR92 pKa = 11.84TGTTEE97 pKa = 4.22LDD99 pKa = 3.6FMGAGPWTLYY109 pKa = 10.07PSSARR114 pKa = 11.84ALYY117 pKa = 10.02SSKK120 pKa = 10.59EE121 pKa = 4.27STVHH125 pKa = 6.82LVDD128 pKa = 3.38EE129 pKa = 4.7AQRR132 pKa = 11.84RR133 pKa = 11.84MIMSKK138 pKa = 9.89CPTMAGPTSTGTYY151 pKa = 7.68MAAIHH156 pKa = 6.28EE157 pKa = 4.49RR158 pKa = 11.84LSTPSDD164 pKa = 3.29LRR166 pKa = 11.84HH167 pKa = 6.52LFTKK171 pKa = 9.88MVLYY175 pKa = 10.65LYY177 pKa = 9.94DD178 pKa = 3.32YY179 pKa = 9.99TLAVVSKK186 pKa = 10.44DD187 pKa = 3.59NTMHH191 pKa = 6.37TADD194 pKa = 5.76DD195 pKa = 3.47ISFDD199 pKa = 4.2LRR201 pKa = 11.84PHH203 pKa = 6.74LPSDD207 pKa = 3.56ALLATVINHH216 pKa = 6.67DD217 pKa = 3.83MVIDD221 pKa = 4.23GDD223 pKa = 4.43LFTPEE228 pKa = 4.52QISLLCLAGQQYY240 pKa = 10.23PSVWYY245 pKa = 10.03AGEE248 pKa = 3.98GNIYY252 pKa = 10.31NSCNMVADD260 pKa = 4.24DD261 pKa = 4.62LVVVSSGRR269 pKa = 11.84LTTDD273 pKa = 3.3SAFTWGSPDD282 pKa = 3.46KK283 pKa = 10.03LYY285 pKa = 11.74NMMWTIAQKK294 pKa = 11.04LNGVSCLMYY303 pKa = 10.62ALEE306 pKa = 4.5SMRR309 pKa = 11.84GKK311 pKa = 10.46CKK313 pKa = 9.91MMSDD317 pKa = 3.21IVAKK321 pKa = 9.49TDD323 pKa = 3.41CRR325 pKa = 11.84EE326 pKa = 3.92VNAMIPRR333 pKa = 11.84SYY335 pKa = 11.26CMSTAFGQIRR345 pKa = 11.84EE346 pKa = 4.11KK347 pKa = 10.87QIVVKK352 pKa = 10.0MPGYY356 pKa = 9.76FSTSIGMLSDD366 pKa = 3.81LMYY369 pKa = 11.34GMTFKK374 pKa = 10.92AVASCVAEE382 pKa = 4.01TLGAMGTIVSSSTPRR397 pKa = 11.84TNPTINGLMRR407 pKa = 11.84DD408 pKa = 3.64YY409 pKa = 11.49GLQHH413 pKa = 5.99TNAWDD418 pKa = 3.47NFMLRR423 pKa = 11.84NFEE426 pKa = 4.2MVTRR430 pKa = 11.84RR431 pKa = 11.84PTQWDD436 pKa = 2.82IGQHH440 pKa = 4.05MKK442 pKa = 10.57EE443 pKa = 4.13YY444 pKa = 10.66ALALAEE450 pKa = 4.29HH451 pKa = 6.36VMLGYY456 pKa = 10.44DD457 pKa = 3.06IEE459 pKa = 4.5MPSILLTIPALTAVNTAYY477 pKa = 10.73GLTRR481 pKa = 11.84GWYY484 pKa = 9.93GGGSTLDD491 pKa = 3.28MDD493 pKa = 4.11KK494 pKa = 10.81KK495 pKa = 10.25QRR497 pKa = 11.84KK498 pKa = 8.33EE499 pKa = 3.92STDD502 pKa = 3.38ALCAVGWMCGLRR514 pKa = 11.84QCRR517 pKa = 11.84PQVFRR522 pKa = 11.84NRR524 pKa = 11.84AGKK527 pKa = 8.94KK528 pKa = 7.3QVMVNAAEE536 pKa = 4.0RR537 pKa = 11.84KK538 pKa = 9.23LRR540 pKa = 11.84AEE542 pKa = 4.56AGDD545 pKa = 3.65DD546 pKa = 3.42CRR548 pKa = 11.84IRR550 pKa = 11.84DD551 pKa = 3.56VEE553 pKa = 4.57FWLEE557 pKa = 3.88DD558 pKa = 3.74TPGGRR563 pKa = 11.84VDD565 pKa = 4.04EE566 pKa = 4.82NEE568 pKa = 3.72EE569 pKa = 4.19SAPNLYY575 pKa = 8.59KK576 pKa = 10.56TEE578 pKa = 4.18FSGTKK583 pKa = 9.44CAMVFNYY590 pKa = 9.94EE591 pKa = 3.38MGMWIEE597 pKa = 4.21ARR599 pKa = 11.84QMDD602 pKa = 4.49YY603 pKa = 11.52DD604 pKa = 3.7RR605 pKa = 11.84LKK607 pKa = 11.0RR608 pKa = 11.84EE609 pKa = 4.2TFSGDD614 pKa = 2.95LTKK617 pKa = 10.53KK618 pKa = 9.89EE619 pKa = 4.38RR620 pKa = 11.84YY621 pKa = 8.06TMSKK625 pKa = 9.43VSAMPIHH632 pKa = 6.97WGPPPNHH639 pKa = 6.72KK640 pKa = 10.61AKK642 pKa = 10.91LEE644 pKa = 4.01ASLEE648 pKa = 4.18HH649 pKa = 6.36MKK651 pKa = 10.75SISRR655 pKa = 11.84GNAIVPTRR663 pKa = 11.84EE664 pKa = 4.02PKK666 pKa = 9.85HH667 pKa = 6.69VRR669 pKa = 11.84INSQSMAVVPKK680 pKa = 9.34YY681 pKa = 11.0VKK683 pKa = 10.75DD684 pKa = 3.55GVEE687 pKa = 3.83EE688 pKa = 4.02EE689 pKa = 4.9KK690 pKa = 10.75YY691 pKa = 9.68VHH693 pKa = 5.89YY694 pKa = 9.71EE695 pKa = 3.75RR696 pKa = 11.84PAIEE700 pKa = 4.4EE701 pKa = 4.1GDD703 pKa = 3.63TIRR706 pKa = 11.84FSEE709 pKa = 4.23IDD711 pKa = 3.48VPGDD715 pKa = 3.73GSCGIHH721 pKa = 7.4AMVKK725 pKa = 10.22DD726 pKa = 3.73LTVHH730 pKa = 6.1GRR732 pKa = 11.84LSPHH736 pKa = 6.23EE737 pKa = 3.98AAKK740 pKa = 9.44ATEE743 pKa = 4.38LFSTDD748 pKa = 2.93TASKK752 pKa = 9.99KK753 pKa = 10.2FHH755 pKa = 7.22DD756 pKa = 4.48AAEE759 pKa = 4.33LAAQCQLWGMGMDD772 pKa = 5.87LIDD775 pKa = 4.05KK776 pKa = 10.11GSNRR780 pKa = 11.84VTRR783 pKa = 11.84YY784 pKa = 9.86GPEE787 pKa = 3.82DD788 pKa = 3.44SEE790 pKa = 4.49YY791 pKa = 10.65SITIIRR797 pKa = 11.84DD798 pKa = 3.19GGHH801 pKa = 5.81FRR803 pKa = 11.84AGLIGEE809 pKa = 5.04GANEE813 pKa = 3.8MTVEE817 pKa = 4.06HH818 pKa = 7.08LEE820 pKa = 4.15QQTRR824 pKa = 11.84APEE827 pKa = 3.92EE828 pKa = 3.7FVRR831 pKa = 11.84DD832 pKa = 4.06VKK834 pKa = 11.38SLGSLFGGSPILQQ847 pKa = 3.42
MM1 pKa = 7.24AAEE4 pKa = 4.4ARR6 pKa = 11.84VIDD9 pKa = 4.24YY10 pKa = 8.19MKK12 pKa = 10.71GLPQSYY18 pKa = 6.77QHH20 pKa = 6.8RR21 pKa = 11.84EE22 pKa = 3.69TYY24 pKa = 10.4FEE26 pKa = 4.74QFLEE30 pKa = 4.34QNSEE34 pKa = 4.16RR35 pKa = 11.84LVEE38 pKa = 4.75EE39 pKa = 3.44IDD41 pKa = 3.91GPVVEE46 pKa = 5.52KK47 pKa = 11.11GLISDD52 pKa = 4.37GEE54 pKa = 4.23KK55 pKa = 10.56EE56 pKa = 4.42IIRR59 pKa = 11.84HH60 pKa = 5.96LNVQDD65 pKa = 3.46KK66 pKa = 8.36FTINGFRR73 pKa = 11.84KK74 pKa = 9.47SVKK77 pKa = 8.0ICHH80 pKa = 5.5MVSLGATRR88 pKa = 11.84QTKK91 pKa = 8.55RR92 pKa = 11.84TGTTEE97 pKa = 4.22LDD99 pKa = 3.6FMGAGPWTLYY109 pKa = 10.07PSSARR114 pKa = 11.84ALYY117 pKa = 10.02SSKK120 pKa = 10.59EE121 pKa = 4.27STVHH125 pKa = 6.82LVDD128 pKa = 3.38EE129 pKa = 4.7AQRR132 pKa = 11.84RR133 pKa = 11.84MIMSKK138 pKa = 9.89CPTMAGPTSTGTYY151 pKa = 7.68MAAIHH156 pKa = 6.28EE157 pKa = 4.49RR158 pKa = 11.84LSTPSDD164 pKa = 3.29LRR166 pKa = 11.84HH167 pKa = 6.52LFTKK171 pKa = 9.88MVLYY175 pKa = 10.65LYY177 pKa = 9.94DD178 pKa = 3.32YY179 pKa = 9.99TLAVVSKK186 pKa = 10.44DD187 pKa = 3.59NTMHH191 pKa = 6.37TADD194 pKa = 5.76DD195 pKa = 3.47ISFDD199 pKa = 4.2LRR201 pKa = 11.84PHH203 pKa = 6.74LPSDD207 pKa = 3.56ALLATVINHH216 pKa = 6.67DD217 pKa = 3.83MVIDD221 pKa = 4.23GDD223 pKa = 4.43LFTPEE228 pKa = 4.52QISLLCLAGQQYY240 pKa = 10.23PSVWYY245 pKa = 10.03AGEE248 pKa = 3.98GNIYY252 pKa = 10.31NSCNMVADD260 pKa = 4.24DD261 pKa = 4.62LVVVSSGRR269 pKa = 11.84LTTDD273 pKa = 3.3SAFTWGSPDD282 pKa = 3.46KK283 pKa = 10.03LYY285 pKa = 11.74NMMWTIAQKK294 pKa = 11.04LNGVSCLMYY303 pKa = 10.62ALEE306 pKa = 4.5SMRR309 pKa = 11.84GKK311 pKa = 10.46CKK313 pKa = 9.91MMSDD317 pKa = 3.21IVAKK321 pKa = 9.49TDD323 pKa = 3.41CRR325 pKa = 11.84EE326 pKa = 3.92VNAMIPRR333 pKa = 11.84SYY335 pKa = 11.26CMSTAFGQIRR345 pKa = 11.84EE346 pKa = 4.11KK347 pKa = 10.87QIVVKK352 pKa = 10.0MPGYY356 pKa = 9.76FSTSIGMLSDD366 pKa = 3.81LMYY369 pKa = 11.34GMTFKK374 pKa = 10.92AVASCVAEE382 pKa = 4.01TLGAMGTIVSSSTPRR397 pKa = 11.84TNPTINGLMRR407 pKa = 11.84DD408 pKa = 3.64YY409 pKa = 11.49GLQHH413 pKa = 5.99TNAWDD418 pKa = 3.47NFMLRR423 pKa = 11.84NFEE426 pKa = 4.2MVTRR430 pKa = 11.84RR431 pKa = 11.84PTQWDD436 pKa = 2.82IGQHH440 pKa = 4.05MKK442 pKa = 10.57EE443 pKa = 4.13YY444 pKa = 10.66ALALAEE450 pKa = 4.29HH451 pKa = 6.36VMLGYY456 pKa = 10.44DD457 pKa = 3.06IEE459 pKa = 4.5MPSILLTIPALTAVNTAYY477 pKa = 10.73GLTRR481 pKa = 11.84GWYY484 pKa = 9.93GGGSTLDD491 pKa = 3.28MDD493 pKa = 4.11KK494 pKa = 10.81KK495 pKa = 10.25QRR497 pKa = 11.84KK498 pKa = 8.33EE499 pKa = 3.92STDD502 pKa = 3.38ALCAVGWMCGLRR514 pKa = 11.84QCRR517 pKa = 11.84PQVFRR522 pKa = 11.84NRR524 pKa = 11.84AGKK527 pKa = 8.94KK528 pKa = 7.3QVMVNAAEE536 pKa = 4.0RR537 pKa = 11.84KK538 pKa = 9.23LRR540 pKa = 11.84AEE542 pKa = 4.56AGDD545 pKa = 3.65DD546 pKa = 3.42CRR548 pKa = 11.84IRR550 pKa = 11.84DD551 pKa = 3.56VEE553 pKa = 4.57FWLEE557 pKa = 3.88DD558 pKa = 3.74TPGGRR563 pKa = 11.84VDD565 pKa = 4.04EE566 pKa = 4.82NEE568 pKa = 3.72EE569 pKa = 4.19SAPNLYY575 pKa = 8.59KK576 pKa = 10.56TEE578 pKa = 4.18FSGTKK583 pKa = 9.44CAMVFNYY590 pKa = 9.94EE591 pKa = 3.38MGMWIEE597 pKa = 4.21ARR599 pKa = 11.84QMDD602 pKa = 4.49YY603 pKa = 11.52DD604 pKa = 3.7RR605 pKa = 11.84LKK607 pKa = 11.0RR608 pKa = 11.84EE609 pKa = 4.2TFSGDD614 pKa = 2.95LTKK617 pKa = 10.53KK618 pKa = 9.89EE619 pKa = 4.38RR620 pKa = 11.84YY621 pKa = 8.06TMSKK625 pKa = 9.43VSAMPIHH632 pKa = 6.97WGPPPNHH639 pKa = 6.72KK640 pKa = 10.61AKK642 pKa = 10.91LEE644 pKa = 4.01ASLEE648 pKa = 4.18HH649 pKa = 6.36MKK651 pKa = 10.75SISRR655 pKa = 11.84GNAIVPTRR663 pKa = 11.84EE664 pKa = 4.02PKK666 pKa = 9.85HH667 pKa = 6.69VRR669 pKa = 11.84INSQSMAVVPKK680 pKa = 9.34YY681 pKa = 11.0VKK683 pKa = 10.75DD684 pKa = 3.55GVEE687 pKa = 3.83EE688 pKa = 4.02EE689 pKa = 4.9KK690 pKa = 10.75YY691 pKa = 9.68VHH693 pKa = 5.89YY694 pKa = 9.71EE695 pKa = 3.75RR696 pKa = 11.84PAIEE700 pKa = 4.4EE701 pKa = 4.1GDD703 pKa = 3.63TIRR706 pKa = 11.84FSEE709 pKa = 4.23IDD711 pKa = 3.48VPGDD715 pKa = 3.73GSCGIHH721 pKa = 7.4AMVKK725 pKa = 10.22DD726 pKa = 3.73LTVHH730 pKa = 6.1GRR732 pKa = 11.84LSPHH736 pKa = 6.23EE737 pKa = 3.98AAKK740 pKa = 9.44ATEE743 pKa = 4.38LFSTDD748 pKa = 2.93TASKK752 pKa = 9.99KK753 pKa = 10.2FHH755 pKa = 7.22DD756 pKa = 4.48AAEE759 pKa = 4.33LAAQCQLWGMGMDD772 pKa = 5.87LIDD775 pKa = 4.05KK776 pKa = 10.11GSNRR780 pKa = 11.84VTRR783 pKa = 11.84YY784 pKa = 9.86GPEE787 pKa = 3.82DD788 pKa = 3.44SEE790 pKa = 4.49YY791 pKa = 10.65SITIIRR797 pKa = 11.84DD798 pKa = 3.19GGHH801 pKa = 5.81FRR803 pKa = 11.84AGLIGEE809 pKa = 5.04GANEE813 pKa = 3.8MTVEE817 pKa = 4.06HH818 pKa = 7.08LEE820 pKa = 4.15QQTRR824 pKa = 11.84APEE827 pKa = 3.92EE828 pKa = 3.7FVRR831 pKa = 11.84DD832 pKa = 4.06VKK834 pKa = 11.38SLGSLFGGSPILQQ847 pKa = 3.42
Molecular weight: 94.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8JVC2|RDRP_PCVC RNA-directed RNA polymerase OS=Penicillium chrysogenum virus (isolate Caston/2003) OX=654932 GN=p1 PE=4 SV=1
MM1 pKa = 7.7AAPVLYY7 pKa = 10.52GGAGGTATGPGDD19 pKa = 3.44MRR21 pKa = 11.84RR22 pKa = 11.84SLMHH26 pKa = 6.44EE27 pKa = 4.25KK28 pKa = 10.35KK29 pKa = 10.2QVFAEE34 pKa = 4.16LRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 4.05AQALRR43 pKa = 11.84VAKK46 pKa = 9.6EE47 pKa = 3.81ARR49 pKa = 11.84GKK51 pKa = 9.94MSVWDD56 pKa = 3.73PSTRR60 pKa = 11.84EE61 pKa = 3.49GARR64 pKa = 11.84GYY66 pKa = 9.98RR67 pKa = 11.84EE68 pKa = 3.61KK69 pKa = 10.82VVRR72 pKa = 11.84FGRR75 pKa = 11.84QIASLLQYY83 pKa = 9.67FEE85 pKa = 5.33NMHH88 pKa = 6.43SPALDD93 pKa = 4.05IIACDD98 pKa = 3.61KK99 pKa = 10.96FLLKK103 pKa = 10.4YY104 pKa = 10.25QIYY107 pKa = 10.69GDD109 pKa = 3.91IDD111 pKa = 3.75RR112 pKa = 11.84DD113 pKa = 3.56PAFGEE118 pKa = 3.91NTMTAEE124 pKa = 4.73VPVVWDD130 pKa = 3.31KK131 pKa = 12.04CEE133 pKa = 4.24VEE135 pKa = 4.4VKK137 pKa = 10.38LYY139 pKa = 10.69AGPLQKK145 pKa = 10.84LMSRR149 pKa = 11.84AKK151 pKa = 10.29LVGAARR157 pKa = 11.84EE158 pKa = 4.54GIPNRR163 pKa = 11.84NDD165 pKa = 2.98VAKK168 pKa = 9.95STGWNQDD175 pKa = 3.23QVQKK179 pKa = 11.0FPDD182 pKa = 3.4NRR184 pKa = 11.84MDD186 pKa = 3.8SLISLLEE193 pKa = 3.85QMQTGQSKK201 pKa = 8.36LTRR204 pKa = 11.84LVKK207 pKa = 10.65GFLILLEE214 pKa = 3.88MAEE217 pKa = 4.0RR218 pKa = 11.84KK219 pKa = 9.51EE220 pKa = 4.04VDD222 pKa = 4.05FHH224 pKa = 7.73VGNHH228 pKa = 4.56IHH230 pKa = 5.23VTYY233 pKa = 10.51AIAPVCDD240 pKa = 4.3SYY242 pKa = 12.03DD243 pKa = 3.62LPGRR247 pKa = 11.84CYY249 pKa = 10.96VFNSKK254 pKa = 8.49PTSEE258 pKa = 3.76AHH260 pKa = 6.17AAVLLAMCRR269 pKa = 11.84EE270 pKa = 4.1YY271 pKa = 10.51PPPQFASHH279 pKa = 5.98VSVPADD285 pKa = 3.53AEE287 pKa = 4.39DD288 pKa = 3.6VCIVSQGRR296 pKa = 11.84QIQPGSAVTLNPGLVYY312 pKa = 10.76SSILTYY318 pKa = 11.49AMDD321 pKa = 4.19TSCTDD326 pKa = 4.28LLQEE330 pKa = 4.33AQIIACSLQEE340 pKa = 3.53NRR342 pKa = 11.84YY343 pKa = 8.48FSRR346 pKa = 11.84IGLPTVVSLYY356 pKa = 11.21DD357 pKa = 3.44LMVPAFIAQNSALEE371 pKa = 4.11GARR374 pKa = 11.84LSGDD378 pKa = 3.11LSKK381 pKa = 11.24AVGRR385 pKa = 11.84VHH387 pKa = 6.94QMLGMVAAKK396 pKa = 10.23DD397 pKa = 3.88IISATHH403 pKa = 4.66MQSRR407 pKa = 11.84TGFDD411 pKa = 3.32PSHH414 pKa = 7.51GIRR417 pKa = 11.84QYY419 pKa = 11.54LNSNSRR425 pKa = 11.84LVTQMASKK433 pKa = 9.23LTGIGLFDD441 pKa = 4.12ATPQMRR447 pKa = 11.84IFSEE451 pKa = 4.18MDD453 pKa = 2.86TADD456 pKa = 3.91YY457 pKa = 11.52ADD459 pKa = 4.38MLHH462 pKa = 5.81LTIFEE467 pKa = 4.43GLWLVQDD474 pKa = 4.92ASVCTDD480 pKa = 3.24NGPISFLVNGEE491 pKa = 4.28KK492 pKa = 10.71LLSADD497 pKa = 3.57RR498 pKa = 11.84AGYY501 pKa = 10.0DD502 pKa = 3.29VLVEE506 pKa = 4.22EE507 pKa = 4.84LTLANIRR514 pKa = 11.84IEE516 pKa = 4.04HH517 pKa = 6.44HH518 pKa = 6.52KK519 pKa = 10.08MPTGAFTTRR528 pKa = 11.84WVAAKK533 pKa = 9.85RR534 pKa = 11.84DD535 pKa = 3.59SALRR539 pKa = 11.84LTPRR543 pKa = 11.84SRR545 pKa = 11.84TAHH548 pKa = 5.67RR549 pKa = 11.84VDD551 pKa = 3.56MVRR554 pKa = 11.84EE555 pKa = 4.04CDD557 pKa = 4.52FNPTMNLKK565 pKa = 10.35AAGPKK570 pKa = 9.93ARR572 pKa = 11.84LRR574 pKa = 11.84GSGVKK579 pKa = 9.52SRR581 pKa = 11.84RR582 pKa = 11.84RR583 pKa = 11.84VSEE586 pKa = 3.93VPLAHH591 pKa = 6.81VFRR594 pKa = 11.84SPPRR598 pKa = 11.84RR599 pKa = 11.84EE600 pKa = 4.09STTTTDD606 pKa = 5.2DD607 pKa = 3.38SPRR610 pKa = 11.84WLTRR614 pKa = 11.84EE615 pKa = 4.27GPQLTRR621 pKa = 11.84RR622 pKa = 11.84VPIIDD627 pKa = 3.98EE628 pKa = 4.35PPAYY632 pKa = 9.82EE633 pKa = 4.27SGRR636 pKa = 11.84SSSPVTSSISEE647 pKa = 3.97GTSQHH652 pKa = 5.41EE653 pKa = 4.35EE654 pKa = 3.86EE655 pKa = 5.02MGLFDD660 pKa = 5.81AEE662 pKa = 4.47EE663 pKa = 4.97LPMQQTVIATEE674 pKa = 3.56ARR676 pKa = 11.84RR677 pKa = 11.84RR678 pKa = 11.84LGRR681 pKa = 11.84GTLEE685 pKa = 4.83RR686 pKa = 11.84IQEE689 pKa = 4.15AALEE693 pKa = 4.28GQVAQGEE700 pKa = 4.59VTAEE704 pKa = 3.68KK705 pKa = 10.46NRR707 pKa = 11.84RR708 pKa = 11.84IEE710 pKa = 4.09AMLSARR716 pKa = 11.84DD717 pKa = 3.58PQFTGRR723 pKa = 11.84EE724 pKa = 4.2QITKK728 pKa = 8.74MLSDD732 pKa = 3.68GGLGVRR738 pKa = 11.84EE739 pKa = 3.84RR740 pKa = 11.84EE741 pKa = 3.8EE742 pKa = 3.74WLEE745 pKa = 3.96LVDD748 pKa = 3.61KK749 pKa = 10.3TVGVKK754 pKa = 9.91GLKK757 pKa = 8.54EE758 pKa = 3.81VRR760 pKa = 11.84SIDD763 pKa = 3.7GIRR766 pKa = 11.84RR767 pKa = 11.84HH768 pKa = 5.5LEE770 pKa = 3.71EE771 pKa = 4.71YY772 pKa = 10.87GEE774 pKa = 4.3RR775 pKa = 11.84EE776 pKa = 3.81GFAVVRR782 pKa = 11.84TLLSGNSKK790 pKa = 9.74HH791 pKa = 6.25VRR793 pKa = 11.84RR794 pKa = 11.84INQLIRR800 pKa = 11.84EE801 pKa = 4.45SNPSAFEE808 pKa = 4.16TEE810 pKa = 3.93ASRR813 pKa = 11.84MRR815 pKa = 11.84RR816 pKa = 11.84LRR818 pKa = 11.84ADD820 pKa = 3.02WDD822 pKa = 3.69GDD824 pKa = 3.73AGSAPVNALHH834 pKa = 6.79FVGNSPGWKK843 pKa = 9.09RR844 pKa = 11.84WLEE847 pKa = 4.04NNNIPSDD854 pKa = 3.49IQVAGKK860 pKa = 10.47KK861 pKa = 9.93RR862 pKa = 11.84MCSYY866 pKa = 10.25LAEE869 pKa = 4.29VLSHH873 pKa = 6.67GNLKK877 pKa = 10.79LSDD880 pKa = 3.43ATKK883 pKa = 10.54LGRR886 pKa = 11.84LVEE889 pKa = 4.32GTSLDD894 pKa = 3.74LFPPQLSSEE903 pKa = 4.22EE904 pKa = 4.17FSTCSEE910 pKa = 3.94ATLAWRR916 pKa = 11.84NAPSSLGVRR925 pKa = 11.84PFAQEE930 pKa = 3.62DD931 pKa = 4.09SRR933 pKa = 11.84WLVMAATCGGGSFGIGKK950 pKa = 9.71LKK952 pKa = 10.54SLCKK956 pKa = 10.02EE957 pKa = 3.72FSVPKK962 pKa = 9.77EE963 pKa = 3.65LRR965 pKa = 11.84DD966 pKa = 3.7ALRR969 pKa = 11.84VKK971 pKa = 10.86YY972 pKa = 10.54GLFGGKK978 pKa = 9.8DD979 pKa = 3.26SLEE982 pKa = 4.0
MM1 pKa = 7.7AAPVLYY7 pKa = 10.52GGAGGTATGPGDD19 pKa = 3.44MRR21 pKa = 11.84RR22 pKa = 11.84SLMHH26 pKa = 6.44EE27 pKa = 4.25KK28 pKa = 10.35KK29 pKa = 10.2QVFAEE34 pKa = 4.16LRR36 pKa = 11.84RR37 pKa = 11.84EE38 pKa = 4.05AQALRR43 pKa = 11.84VAKK46 pKa = 9.6EE47 pKa = 3.81ARR49 pKa = 11.84GKK51 pKa = 9.94MSVWDD56 pKa = 3.73PSTRR60 pKa = 11.84EE61 pKa = 3.49GARR64 pKa = 11.84GYY66 pKa = 9.98RR67 pKa = 11.84EE68 pKa = 3.61KK69 pKa = 10.82VVRR72 pKa = 11.84FGRR75 pKa = 11.84QIASLLQYY83 pKa = 9.67FEE85 pKa = 5.33NMHH88 pKa = 6.43SPALDD93 pKa = 4.05IIACDD98 pKa = 3.61KK99 pKa = 10.96FLLKK103 pKa = 10.4YY104 pKa = 10.25QIYY107 pKa = 10.69GDD109 pKa = 3.91IDD111 pKa = 3.75RR112 pKa = 11.84DD113 pKa = 3.56PAFGEE118 pKa = 3.91NTMTAEE124 pKa = 4.73VPVVWDD130 pKa = 3.31KK131 pKa = 12.04CEE133 pKa = 4.24VEE135 pKa = 4.4VKK137 pKa = 10.38LYY139 pKa = 10.69AGPLQKK145 pKa = 10.84LMSRR149 pKa = 11.84AKK151 pKa = 10.29LVGAARR157 pKa = 11.84EE158 pKa = 4.54GIPNRR163 pKa = 11.84NDD165 pKa = 2.98VAKK168 pKa = 9.95STGWNQDD175 pKa = 3.23QVQKK179 pKa = 11.0FPDD182 pKa = 3.4NRR184 pKa = 11.84MDD186 pKa = 3.8SLISLLEE193 pKa = 3.85QMQTGQSKK201 pKa = 8.36LTRR204 pKa = 11.84LVKK207 pKa = 10.65GFLILLEE214 pKa = 3.88MAEE217 pKa = 4.0RR218 pKa = 11.84KK219 pKa = 9.51EE220 pKa = 4.04VDD222 pKa = 4.05FHH224 pKa = 7.73VGNHH228 pKa = 4.56IHH230 pKa = 5.23VTYY233 pKa = 10.51AIAPVCDD240 pKa = 4.3SYY242 pKa = 12.03DD243 pKa = 3.62LPGRR247 pKa = 11.84CYY249 pKa = 10.96VFNSKK254 pKa = 8.49PTSEE258 pKa = 3.76AHH260 pKa = 6.17AAVLLAMCRR269 pKa = 11.84EE270 pKa = 4.1YY271 pKa = 10.51PPPQFASHH279 pKa = 5.98VSVPADD285 pKa = 3.53AEE287 pKa = 4.39DD288 pKa = 3.6VCIVSQGRR296 pKa = 11.84QIQPGSAVTLNPGLVYY312 pKa = 10.76SSILTYY318 pKa = 11.49AMDD321 pKa = 4.19TSCTDD326 pKa = 4.28LLQEE330 pKa = 4.33AQIIACSLQEE340 pKa = 3.53NRR342 pKa = 11.84YY343 pKa = 8.48FSRR346 pKa = 11.84IGLPTVVSLYY356 pKa = 11.21DD357 pKa = 3.44LMVPAFIAQNSALEE371 pKa = 4.11GARR374 pKa = 11.84LSGDD378 pKa = 3.11LSKK381 pKa = 11.24AVGRR385 pKa = 11.84VHH387 pKa = 6.94QMLGMVAAKK396 pKa = 10.23DD397 pKa = 3.88IISATHH403 pKa = 4.66MQSRR407 pKa = 11.84TGFDD411 pKa = 3.32PSHH414 pKa = 7.51GIRR417 pKa = 11.84QYY419 pKa = 11.54LNSNSRR425 pKa = 11.84LVTQMASKK433 pKa = 9.23LTGIGLFDD441 pKa = 4.12ATPQMRR447 pKa = 11.84IFSEE451 pKa = 4.18MDD453 pKa = 2.86TADD456 pKa = 3.91YY457 pKa = 11.52ADD459 pKa = 4.38MLHH462 pKa = 5.81LTIFEE467 pKa = 4.43GLWLVQDD474 pKa = 4.92ASVCTDD480 pKa = 3.24NGPISFLVNGEE491 pKa = 4.28KK492 pKa = 10.71LLSADD497 pKa = 3.57RR498 pKa = 11.84AGYY501 pKa = 10.0DD502 pKa = 3.29VLVEE506 pKa = 4.22EE507 pKa = 4.84LTLANIRR514 pKa = 11.84IEE516 pKa = 4.04HH517 pKa = 6.44HH518 pKa = 6.52KK519 pKa = 10.08MPTGAFTTRR528 pKa = 11.84WVAAKK533 pKa = 9.85RR534 pKa = 11.84DD535 pKa = 3.59SALRR539 pKa = 11.84LTPRR543 pKa = 11.84SRR545 pKa = 11.84TAHH548 pKa = 5.67RR549 pKa = 11.84VDD551 pKa = 3.56MVRR554 pKa = 11.84EE555 pKa = 4.04CDD557 pKa = 4.52FNPTMNLKK565 pKa = 10.35AAGPKK570 pKa = 9.93ARR572 pKa = 11.84LRR574 pKa = 11.84GSGVKK579 pKa = 9.52SRR581 pKa = 11.84RR582 pKa = 11.84RR583 pKa = 11.84VSEE586 pKa = 3.93VPLAHH591 pKa = 6.81VFRR594 pKa = 11.84SPPRR598 pKa = 11.84RR599 pKa = 11.84EE600 pKa = 4.09STTTTDD606 pKa = 5.2DD607 pKa = 3.38SPRR610 pKa = 11.84WLTRR614 pKa = 11.84EE615 pKa = 4.27GPQLTRR621 pKa = 11.84RR622 pKa = 11.84VPIIDD627 pKa = 3.98EE628 pKa = 4.35PPAYY632 pKa = 9.82EE633 pKa = 4.27SGRR636 pKa = 11.84SSSPVTSSISEE647 pKa = 3.97GTSQHH652 pKa = 5.41EE653 pKa = 4.35EE654 pKa = 3.86EE655 pKa = 5.02MGLFDD660 pKa = 5.81AEE662 pKa = 4.47EE663 pKa = 4.97LPMQQTVIATEE674 pKa = 3.56ARR676 pKa = 11.84RR677 pKa = 11.84RR678 pKa = 11.84LGRR681 pKa = 11.84GTLEE685 pKa = 4.83RR686 pKa = 11.84IQEE689 pKa = 4.15AALEE693 pKa = 4.28GQVAQGEE700 pKa = 4.59VTAEE704 pKa = 3.68KK705 pKa = 10.46NRR707 pKa = 11.84RR708 pKa = 11.84IEE710 pKa = 4.09AMLSARR716 pKa = 11.84DD717 pKa = 3.58PQFTGRR723 pKa = 11.84EE724 pKa = 4.2QITKK728 pKa = 8.74MLSDD732 pKa = 3.68GGLGVRR738 pKa = 11.84EE739 pKa = 3.84RR740 pKa = 11.84EE741 pKa = 3.8EE742 pKa = 3.74WLEE745 pKa = 3.96LVDD748 pKa = 3.61KK749 pKa = 10.3TVGVKK754 pKa = 9.91GLKK757 pKa = 8.54EE758 pKa = 3.81VRR760 pKa = 11.84SIDD763 pKa = 3.7GIRR766 pKa = 11.84RR767 pKa = 11.84HH768 pKa = 5.5LEE770 pKa = 3.71EE771 pKa = 4.71YY772 pKa = 10.87GEE774 pKa = 4.3RR775 pKa = 11.84EE776 pKa = 3.81GFAVVRR782 pKa = 11.84TLLSGNSKK790 pKa = 9.74HH791 pKa = 6.25VRR793 pKa = 11.84RR794 pKa = 11.84INQLIRR800 pKa = 11.84EE801 pKa = 4.45SNPSAFEE808 pKa = 4.16TEE810 pKa = 3.93ASRR813 pKa = 11.84MRR815 pKa = 11.84RR816 pKa = 11.84LRR818 pKa = 11.84ADD820 pKa = 3.02WDD822 pKa = 3.69GDD824 pKa = 3.73AGSAPVNALHH834 pKa = 6.79FVGNSPGWKK843 pKa = 9.09RR844 pKa = 11.84WLEE847 pKa = 4.04NNNIPSDD854 pKa = 3.49IQVAGKK860 pKa = 10.47KK861 pKa = 9.93RR862 pKa = 11.84MCSYY866 pKa = 10.25LAEE869 pKa = 4.29VLSHH873 pKa = 6.67GNLKK877 pKa = 10.79LSDD880 pKa = 3.43ATKK883 pKa = 10.54LGRR886 pKa = 11.84LVEE889 pKa = 4.32GTSLDD894 pKa = 3.74LFPPQLSSEE903 pKa = 4.22EE904 pKa = 4.17FSTCSEE910 pKa = 3.94ATLAWRR916 pKa = 11.84NAPSSLGVRR925 pKa = 11.84PFAQEE930 pKa = 3.62DD931 pKa = 4.09SRR933 pKa = 11.84WLVMAATCGGGSFGIGKK950 pKa = 9.71LKK952 pKa = 10.54SLCKK956 pKa = 10.02EE957 pKa = 3.72FSVPKK962 pKa = 9.77EE963 pKa = 3.65LRR965 pKa = 11.84DD966 pKa = 3.7ALRR969 pKa = 11.84VKK971 pKa = 10.86YY972 pKa = 10.54GLFGGKK978 pKa = 9.8DD979 pKa = 3.26SLEE982 pKa = 4.0
Molecular weight: 108.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3858 |
847 |
1117 |
964.5 |
108.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.491 ± 0.493 | 1.788 ± 0.122 |
5.677 ± 0.143 | 7.284 ± 0.22 |
3.37 ± 0.594 | 7.517 ± 0.438 |
2.54 ± 0.15 | 4.303 ± 0.152 |
5.21 ± 0.252 | 9.253 ± 0.505 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.733 ± 0.508 | 3.344 ± 0.338 |
4.044 ± 0.331 | 3.24 ± 0.236 |
7.102 ± 0.408 | 6.713 ± 0.418 |
5.469 ± 0.337 | 7.154 ± 0.47 |
1.788 ± 0.218 | 2.981 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
