
Little cherry virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Ampelovirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
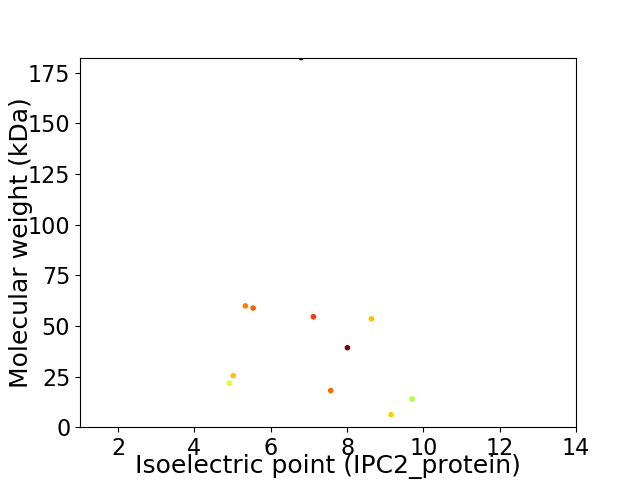
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
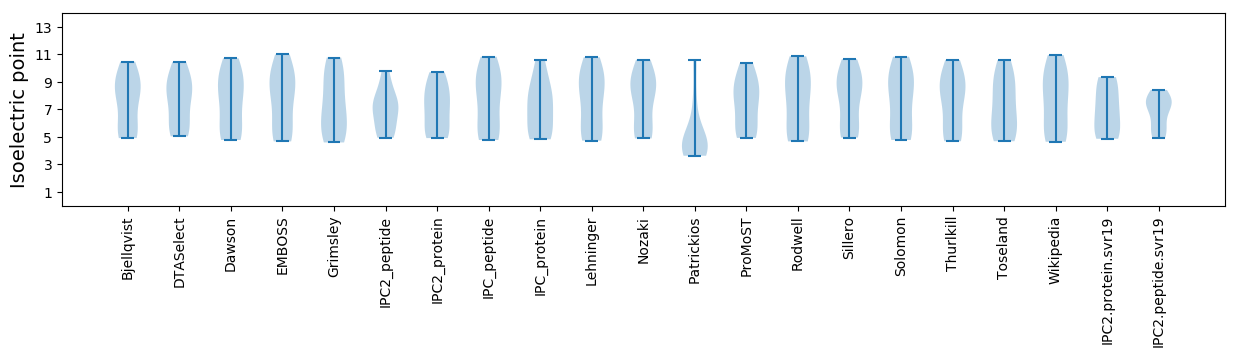
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7T732|Q7T732_9CLOS p53 OS=Little cherry virus 2 OX=154339 PE=4 SV=1
MM1 pKa = 7.14LTLGACLEE9 pKa = 4.27LNFDD13 pKa = 4.21KK14 pKa = 11.21GLTFHH19 pKa = 6.9SRR21 pKa = 11.84YY22 pKa = 9.51KK23 pKa = 10.52RR24 pKa = 11.84SNFLCGSKK32 pKa = 10.25QRR34 pKa = 11.84GDD36 pKa = 5.15APDD39 pKa = 3.51FWYY42 pKa = 10.46EE43 pKa = 3.91KK44 pKa = 10.23IRR46 pKa = 11.84AHH48 pKa = 6.45EE49 pKa = 4.08FDD51 pKa = 3.61EE52 pKa = 5.8VYY54 pKa = 9.27ITDD57 pKa = 3.69YY58 pKa = 10.27AKK60 pKa = 10.76CMSVFEE66 pKa = 4.51PVVVSQPEE74 pKa = 4.04LSKK77 pKa = 10.93LSEE80 pKa = 4.05MSDD83 pKa = 3.47LSLKK87 pKa = 9.76DD88 pKa = 3.29TLLPDD93 pKa = 3.57TDD95 pKa = 4.74LKK97 pKa = 10.42LTYY100 pKa = 10.78GNLEE104 pKa = 4.06LPIKK108 pKa = 10.51KK109 pKa = 9.93LVADD113 pKa = 4.08QLDD116 pKa = 3.76TFDD119 pKa = 4.65NEE121 pKa = 3.77EE122 pKa = 3.69EE123 pKa = 4.79RR124 pKa = 11.84YY125 pKa = 10.5VDD127 pKa = 4.41FSFVGKK133 pKa = 10.24QGEE136 pKa = 4.04KK137 pKa = 10.04HH138 pKa = 5.39YY139 pKa = 11.11FGVICSNQRR148 pKa = 11.84LMTEE152 pKa = 3.51IDD154 pKa = 3.19EE155 pKa = 5.32KK156 pKa = 11.26LFKK159 pKa = 10.43DD160 pKa = 3.89YY161 pKa = 9.09KK162 pKa = 9.37TLVNKK167 pKa = 10.15LVEE170 pKa = 4.15LTEE173 pKa = 4.09EE174 pKa = 4.47KK175 pKa = 10.51IGDD178 pKa = 3.76KK179 pKa = 10.89NLLLFTLTT187 pKa = 3.91
MM1 pKa = 7.14LTLGACLEE9 pKa = 4.27LNFDD13 pKa = 4.21KK14 pKa = 11.21GLTFHH19 pKa = 6.9SRR21 pKa = 11.84YY22 pKa = 9.51KK23 pKa = 10.52RR24 pKa = 11.84SNFLCGSKK32 pKa = 10.25QRR34 pKa = 11.84GDD36 pKa = 5.15APDD39 pKa = 3.51FWYY42 pKa = 10.46EE43 pKa = 3.91KK44 pKa = 10.23IRR46 pKa = 11.84AHH48 pKa = 6.45EE49 pKa = 4.08FDD51 pKa = 3.61EE52 pKa = 5.8VYY54 pKa = 9.27ITDD57 pKa = 3.69YY58 pKa = 10.27AKK60 pKa = 10.76CMSVFEE66 pKa = 4.51PVVVSQPEE74 pKa = 4.04LSKK77 pKa = 10.93LSEE80 pKa = 4.05MSDD83 pKa = 3.47LSLKK87 pKa = 9.76DD88 pKa = 3.29TLLPDD93 pKa = 3.57TDD95 pKa = 4.74LKK97 pKa = 10.42LTYY100 pKa = 10.78GNLEE104 pKa = 4.06LPIKK108 pKa = 10.51KK109 pKa = 9.93LVADD113 pKa = 4.08QLDD116 pKa = 3.76TFDD119 pKa = 4.65NEE121 pKa = 3.77EE122 pKa = 3.69EE123 pKa = 4.79RR124 pKa = 11.84YY125 pKa = 10.5VDD127 pKa = 4.41FSFVGKK133 pKa = 10.24QGEE136 pKa = 4.04KK137 pKa = 10.04HH138 pKa = 5.39YY139 pKa = 11.11FGVICSNQRR148 pKa = 11.84LMTEE152 pKa = 3.51IDD154 pKa = 3.19EE155 pKa = 5.32KK156 pKa = 11.26LFKK159 pKa = 10.43DD160 pKa = 3.89YY161 pKa = 9.09KK162 pKa = 9.37TLVNKK167 pKa = 10.15LVEE170 pKa = 4.15LTEE173 pKa = 4.09EE174 pKa = 4.47KK175 pKa = 10.51IGDD178 pKa = 3.76KK179 pKa = 10.89NLLLFTLTT187 pKa = 3.91
Molecular weight: 21.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7T737|Q7T737_9CLOS RNA-dependent RNA polymerase OS=Little cherry virus 2 OX=154339 PE=4 SV=1
LL1 pKa = 7.48VPHH4 pKa = 7.02ACSTKK9 pKa = 9.56TFPFSLSCSFFHH21 pKa = 7.07QLIQFIKK28 pKa = 10.23LFITIFYY35 pKa = 10.2LGLRR39 pKa = 11.84LASFLSRR46 pKa = 11.84GGLQGPYY53 pKa = 9.5EE54 pKa = 4.2KK55 pKa = 10.24RR56 pKa = 11.84IRR58 pKa = 11.84TPNSEE63 pKa = 4.06VSSEE67 pKa = 3.97FSLDD71 pKa = 3.24LTRR74 pKa = 11.84DD75 pKa = 2.89KK76 pKa = 11.03RR77 pKa = 11.84FRR79 pKa = 11.84EE80 pKa = 4.23LSRR83 pKa = 11.84LPGVVPTMYY92 pKa = 10.71CHH94 pKa = 6.04GCKK97 pKa = 9.73RR98 pKa = 11.84APLVARR104 pKa = 11.84TRR106 pKa = 11.84QRR108 pKa = 11.84YY109 pKa = 4.6EE110 pKa = 3.24ARR112 pKa = 11.84VRR114 pKa = 11.84VLLVAWSS121 pKa = 3.4
LL1 pKa = 7.48VPHH4 pKa = 7.02ACSTKK9 pKa = 9.56TFPFSLSCSFFHH21 pKa = 7.07QLIQFIKK28 pKa = 10.23LFITIFYY35 pKa = 10.2LGLRR39 pKa = 11.84LASFLSRR46 pKa = 11.84GGLQGPYY53 pKa = 9.5EE54 pKa = 4.2KK55 pKa = 10.24RR56 pKa = 11.84IRR58 pKa = 11.84TPNSEE63 pKa = 4.06VSSEE67 pKa = 3.97FSLDD71 pKa = 3.24LTRR74 pKa = 11.84DD75 pKa = 2.89KK76 pKa = 11.03RR77 pKa = 11.84FRR79 pKa = 11.84EE80 pKa = 4.23LSRR83 pKa = 11.84LPGVVPTMYY92 pKa = 10.71CHH94 pKa = 6.04GCKK97 pKa = 9.73RR98 pKa = 11.84APLVARR104 pKa = 11.84TRR106 pKa = 11.84QRR108 pKa = 11.84YY109 pKa = 4.6EE110 pKa = 3.24ARR112 pKa = 11.84VRR114 pKa = 11.84VLLVAWSS121 pKa = 3.4
Molecular weight: 13.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4744 |
56 |
1640 |
431.3 |
48.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.08 ± 0.354 | 1.939 ± 0.219 |
6.64 ± 0.474 | 5.839 ± 0.571 |
4.869 ± 0.411 | 5.712 ± 0.468 |
1.623 ± 0.148 | 4.911 ± 0.362 |
7.357 ± 0.381 | 9.823 ± 0.563 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.045 ± 0.231 | 5.207 ± 0.229 |
3.731 ± 0.48 | 2.319 ± 0.224 |
5.312 ± 0.478 | 9.212 ± 0.6 |
5.965 ± 0.38 | 7.947 ± 0.676 |
0.822 ± 0.126 | 3.647 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
