
Arboretum almendravirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Almendravirus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
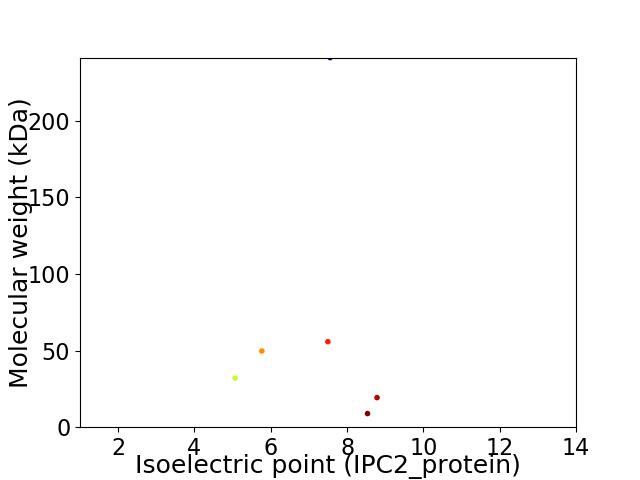
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X4QQM2|X4QQM2_9RHAB SH protein OS=Arboretum almendravirus OX=1972683 PE=4 SV=1
MM1 pKa = 7.6SNPKK5 pKa = 9.93VCDD8 pKa = 3.53DD9 pKa = 4.29LFNSRR14 pKa = 11.84RR15 pKa = 11.84MLAQIDD21 pKa = 3.78VSAFDD26 pKa = 3.26ISKK29 pKa = 10.57QISEE33 pKa = 4.92DD34 pKa = 3.57YY35 pKa = 9.26GTTRR39 pKa = 11.84GTDD42 pKa = 3.25EE43 pKa = 5.29LNDD46 pKa = 3.25IEE48 pKa = 4.75YY49 pKa = 10.98EE50 pKa = 4.05EE51 pKa = 4.82NEE53 pKa = 4.19EE54 pKa = 4.97KK55 pKa = 10.62KK56 pKa = 10.96GLEE59 pKa = 3.73NDD61 pKa = 3.22KK62 pKa = 10.51TKK64 pKa = 10.18TLTGDD69 pKa = 3.86EE70 pKa = 4.25IMTPNIFSRR79 pKa = 11.84MINTPLEE86 pKa = 4.21KK87 pKa = 10.8ANLEE91 pKa = 4.0LSEE94 pKa = 4.47TKK96 pKa = 10.3CRR98 pKa = 11.84NNKK101 pKa = 9.13PCVEE105 pKa = 3.94PRR107 pKa = 11.84QVTFNTGRR115 pKa = 11.84STEE118 pKa = 4.27SNDD121 pKa = 3.24GNKK124 pKa = 10.02SYY126 pKa = 11.55EE127 pKa = 4.04EE128 pKa = 4.91GYY130 pKa = 10.82LRR132 pKa = 11.84AIWDD136 pKa = 3.68INNLLSKK143 pKa = 10.68EE144 pKa = 4.01NFKK147 pKa = 10.47MEE149 pKa = 3.87ITRR152 pKa = 11.84DD153 pKa = 3.67PKK155 pKa = 11.12GSLTIKK161 pKa = 9.15KK162 pKa = 8.43TNDD165 pKa = 2.92SGNVEE170 pKa = 4.07KK171 pKa = 10.79CIEE174 pKa = 4.01LAKK177 pKa = 10.4SDD179 pKa = 4.32EE180 pKa = 4.49SSCSYY185 pKa = 9.16KK186 pKa = 10.44TEE188 pKa = 3.7IDD190 pKa = 3.22QDD192 pKa = 3.3ISFTDD197 pKa = 3.71SEE199 pKa = 5.0SCGVYY204 pKa = 10.23PPLHH208 pKa = 6.44YY209 pKa = 11.11DD210 pKa = 3.54LMFGGKK216 pKa = 8.97VEE218 pKa = 4.32INVKK222 pKa = 10.34VLVEE226 pKa = 4.04EE227 pKa = 4.67LKK229 pKa = 10.97SSNMDD234 pKa = 3.36KK235 pKa = 11.12DD236 pKa = 3.59DD237 pKa = 4.27LKK239 pKa = 11.05IVIQLIDD246 pKa = 3.62NNLEE250 pKa = 3.83NKK252 pKa = 8.75NAKK255 pKa = 10.17DD256 pKa = 2.83ILKK259 pKa = 10.03YY260 pKa = 10.13IKK262 pKa = 10.02HH263 pKa = 5.77SKK265 pKa = 9.79QYY267 pKa = 10.99GKK269 pKa = 9.79IVYY272 pKa = 9.21QVARR276 pKa = 11.84KK277 pKa = 9.82YY278 pKa = 10.79KK279 pKa = 9.45II280 pKa = 3.25
MM1 pKa = 7.6SNPKK5 pKa = 9.93VCDD8 pKa = 3.53DD9 pKa = 4.29LFNSRR14 pKa = 11.84RR15 pKa = 11.84MLAQIDD21 pKa = 3.78VSAFDD26 pKa = 3.26ISKK29 pKa = 10.57QISEE33 pKa = 4.92DD34 pKa = 3.57YY35 pKa = 9.26GTTRR39 pKa = 11.84GTDD42 pKa = 3.25EE43 pKa = 5.29LNDD46 pKa = 3.25IEE48 pKa = 4.75YY49 pKa = 10.98EE50 pKa = 4.05EE51 pKa = 4.82NEE53 pKa = 4.19EE54 pKa = 4.97KK55 pKa = 10.62KK56 pKa = 10.96GLEE59 pKa = 3.73NDD61 pKa = 3.22KK62 pKa = 10.51TKK64 pKa = 10.18TLTGDD69 pKa = 3.86EE70 pKa = 4.25IMTPNIFSRR79 pKa = 11.84MINTPLEE86 pKa = 4.21KK87 pKa = 10.8ANLEE91 pKa = 4.0LSEE94 pKa = 4.47TKK96 pKa = 10.3CRR98 pKa = 11.84NNKK101 pKa = 9.13PCVEE105 pKa = 3.94PRR107 pKa = 11.84QVTFNTGRR115 pKa = 11.84STEE118 pKa = 4.27SNDD121 pKa = 3.24GNKK124 pKa = 10.02SYY126 pKa = 11.55EE127 pKa = 4.04EE128 pKa = 4.91GYY130 pKa = 10.82LRR132 pKa = 11.84AIWDD136 pKa = 3.68INNLLSKK143 pKa = 10.68EE144 pKa = 4.01NFKK147 pKa = 10.47MEE149 pKa = 3.87ITRR152 pKa = 11.84DD153 pKa = 3.67PKK155 pKa = 11.12GSLTIKK161 pKa = 9.15KK162 pKa = 8.43TNDD165 pKa = 2.92SGNVEE170 pKa = 4.07KK171 pKa = 10.79CIEE174 pKa = 4.01LAKK177 pKa = 10.4SDD179 pKa = 4.32EE180 pKa = 4.49SSCSYY185 pKa = 9.16KK186 pKa = 10.44TEE188 pKa = 3.7IDD190 pKa = 3.22QDD192 pKa = 3.3ISFTDD197 pKa = 3.71SEE199 pKa = 5.0SCGVYY204 pKa = 10.23PPLHH208 pKa = 6.44YY209 pKa = 11.11DD210 pKa = 3.54LMFGGKK216 pKa = 8.97VEE218 pKa = 4.32INVKK222 pKa = 10.34VLVEE226 pKa = 4.04EE227 pKa = 4.67LKK229 pKa = 10.97SSNMDD234 pKa = 3.36KK235 pKa = 11.12DD236 pKa = 3.59DD237 pKa = 4.27LKK239 pKa = 11.05IVIQLIDD246 pKa = 3.62NNLEE250 pKa = 3.83NKK252 pKa = 8.75NAKK255 pKa = 10.17DD256 pKa = 2.83ILKK259 pKa = 10.03YY260 pKa = 10.13IKK262 pKa = 10.02HH263 pKa = 5.77SKK265 pKa = 9.79QYY267 pKa = 10.99GKK269 pKa = 9.79IVYY272 pKa = 9.21QVARR276 pKa = 11.84KK277 pKa = 9.82YY278 pKa = 10.79KK279 pKa = 9.45II280 pKa = 3.25
Molecular weight: 32.04 kDa
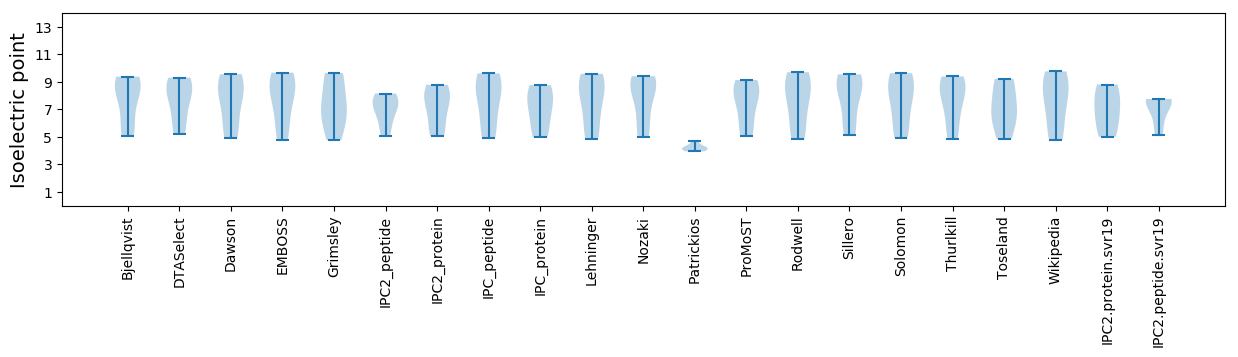
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X4R558|X4R558_9RHAB G protein OS=Arboretum almendravirus OX=1972683 PE=4 SV=1
MM1 pKa = 7.67LANQFIMNNKK11 pKa = 9.35YY12 pKa = 9.44VFKK15 pKa = 9.94TFRR18 pKa = 11.84IRR20 pKa = 11.84YY21 pKa = 5.81EE22 pKa = 3.83ASFKK26 pKa = 10.44EE27 pKa = 4.54LPPHH31 pKa = 7.35SDD33 pKa = 3.85FKK35 pKa = 11.06PILDD39 pKa = 4.18IYY41 pKa = 10.48DD42 pKa = 3.75GKK44 pKa = 10.91IKK46 pKa = 10.34FYY48 pKa = 11.04EE49 pKa = 4.11VFTFIINYY57 pKa = 9.4LLHH60 pKa = 6.04LQIRR64 pKa = 11.84NKK66 pKa = 9.69VMRR69 pKa = 11.84SHH71 pKa = 7.75DD72 pKa = 3.89STFVMCFPVDD82 pKa = 3.63EE83 pKa = 4.4TWTYY87 pKa = 11.1RR88 pKa = 11.84RR89 pKa = 11.84DD90 pKa = 3.98DD91 pKa = 3.49IYY93 pKa = 11.59SFHH96 pKa = 7.78KK97 pKa = 10.04DD98 pKa = 2.55IQMVVGCEE106 pKa = 3.84VRR108 pKa = 11.84EE109 pKa = 3.82LRR111 pKa = 11.84LRR113 pKa = 11.84VIIDD117 pKa = 3.38MNTNNLPRR125 pKa = 11.84SRR127 pKa = 11.84LVRR130 pKa = 11.84DD131 pKa = 3.66VLRR134 pKa = 11.84KK135 pKa = 7.78TLSRR139 pKa = 11.84LYY141 pKa = 10.86DD142 pKa = 3.47PAISSKK148 pKa = 9.4MYY150 pKa = 10.47EE151 pKa = 4.03VIKK154 pKa = 10.51RR155 pKa = 11.84QNEE158 pKa = 4.18CC159 pKa = 3.56
MM1 pKa = 7.67LANQFIMNNKK11 pKa = 9.35YY12 pKa = 9.44VFKK15 pKa = 9.94TFRR18 pKa = 11.84IRR20 pKa = 11.84YY21 pKa = 5.81EE22 pKa = 3.83ASFKK26 pKa = 10.44EE27 pKa = 4.54LPPHH31 pKa = 7.35SDD33 pKa = 3.85FKK35 pKa = 11.06PILDD39 pKa = 4.18IYY41 pKa = 10.48DD42 pKa = 3.75GKK44 pKa = 10.91IKK46 pKa = 10.34FYY48 pKa = 11.04EE49 pKa = 4.11VFTFIINYY57 pKa = 9.4LLHH60 pKa = 6.04LQIRR64 pKa = 11.84NKK66 pKa = 9.69VMRR69 pKa = 11.84SHH71 pKa = 7.75DD72 pKa = 3.89STFVMCFPVDD82 pKa = 3.63EE83 pKa = 4.4TWTYY87 pKa = 11.1RR88 pKa = 11.84RR89 pKa = 11.84DD90 pKa = 3.98DD91 pKa = 3.49IYY93 pKa = 11.59SFHH96 pKa = 7.78KK97 pKa = 10.04DD98 pKa = 2.55IQMVVGCEE106 pKa = 3.84VRR108 pKa = 11.84EE109 pKa = 3.82LRR111 pKa = 11.84LRR113 pKa = 11.84VIIDD117 pKa = 3.38MNTNNLPRR125 pKa = 11.84SRR127 pKa = 11.84LVRR130 pKa = 11.84DD131 pKa = 3.66VLRR134 pKa = 11.84KK135 pKa = 7.78TLSRR139 pKa = 11.84LYY141 pKa = 10.86DD142 pKa = 3.47PAISSKK148 pKa = 9.4MYY150 pKa = 10.47EE151 pKa = 4.03VIKK154 pKa = 10.51RR155 pKa = 11.84QNEE158 pKa = 4.18CC159 pKa = 3.56
Molecular weight: 19.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3484 |
73 |
2062 |
580.7 |
67.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.641 ± 0.493 | 1.464 ± 0.332 |
6.544 ± 0.437 | 6.228 ± 0.586 |
4.65 ± 0.497 | 4.277 ± 0.393 |
2.067 ± 0.292 | 8.927 ± 0.293 |
8.266 ± 0.621 | 9.816 ± 0.749 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.408 | 7.836 ± 0.377 |
3.243 ± 0.246 | 2.612 ± 0.278 |
4.908 ± 0.487 | 7.52 ± 0.168 |
4.822 ± 0.231 | 5.138 ± 0.371 |
1.234 ± 0.212 | 5.253 ± 0.347 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
