
Sugarcane striate virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
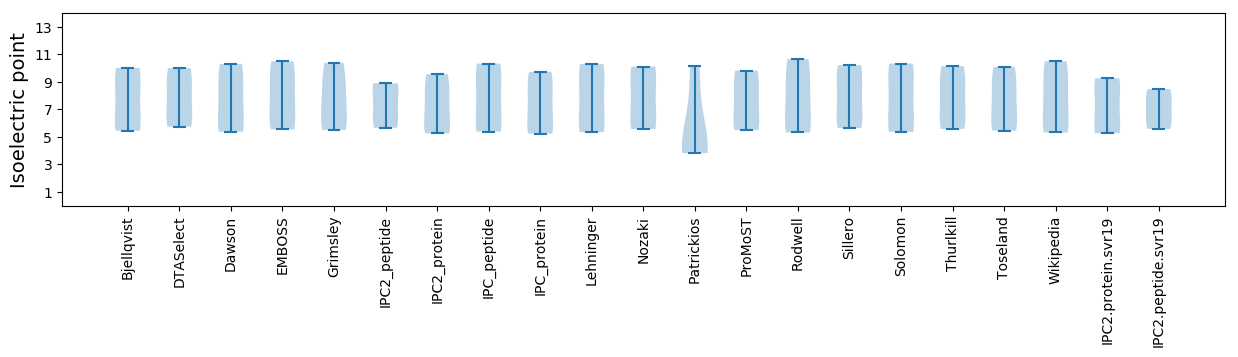
Average proteome isoelectric point is 7.23
Get precalculated fractions of proteins
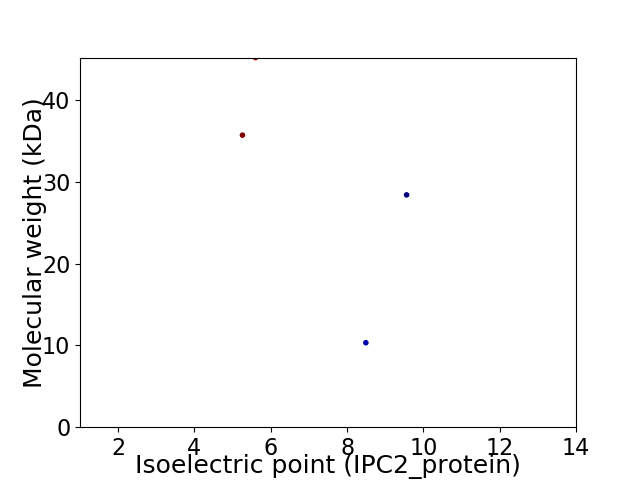
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A218KH22|A0A218KH22_9GEMI Capsid protein OS=Sugarcane striate virus OX=1868659 PE=3 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84TNANAVRR9 pKa = 11.84RR10 pKa = 11.84VEE12 pKa = 4.94FMTTPATTEE21 pKa = 4.01DD22 pKa = 3.79AFQSGTQEE30 pKa = 4.31SSDD33 pKa = 3.85SPSEE37 pKa = 4.44PIHH40 pKa = 5.16THH42 pKa = 3.12QTTWKK47 pKa = 9.41RR48 pKa = 11.84NFEE51 pKa = 4.06FRR53 pKa = 11.84SCNAFLTYY61 pKa = 9.53PRR63 pKa = 11.84CALDD67 pKa = 3.57CEE69 pKa = 4.63YY70 pKa = 11.46VLDD73 pKa = 4.73LPVGSYY79 pKa = 7.89VFRR82 pKa = 11.84WSPIYY87 pKa = 10.88ALVSAEE93 pKa = 3.76NHH95 pKa = 5.26QDD97 pKa = 3.26GGIHH101 pKa = 6.03LHH103 pKa = 6.7CLIQTLGRR111 pKa = 11.84ITTRR115 pKa = 11.84DD116 pKa = 3.27PAFFDD121 pKa = 5.1LDD123 pKa = 3.73NQYY126 pKa = 10.86HH127 pKa = 6.7PNIQAARR134 pKa = 11.84SPDD137 pKa = 3.22NTRR140 pKa = 11.84DD141 pKa = 3.81YY142 pKa = 11.06ILKK145 pKa = 10.47DD146 pKa = 3.85PISHH150 pKa = 5.64VEE152 pKa = 3.81RR153 pKa = 11.84GTFQPRR159 pKa = 11.84RR160 pKa = 11.84GNTRR164 pKa = 11.84NPGDD168 pKa = 3.75RR169 pKa = 11.84GSGSKK174 pKa = 10.26DD175 pKa = 3.09AIMKK179 pKa = 9.31EE180 pKa = 3.93IFRR183 pKa = 11.84TATSKK188 pKa = 10.91EE189 pKa = 3.99DD190 pKa = 3.83YY191 pKa = 10.93LRR193 pKa = 11.84MIQKK197 pKa = 9.61SFPFEE202 pKa = 3.95WAKK205 pKa = 10.73HH206 pKa = 4.91LSQFEE211 pKa = 3.98YY212 pKa = 10.21SANYY216 pKa = 9.33LFPPVPEE223 pKa = 4.46PFNNDD228 pKa = 3.09FPEE231 pKa = 4.38TEE233 pKa = 4.15PDD235 pKa = 4.28LICRR239 pKa = 11.84EE240 pKa = 4.27VLDD243 pKa = 4.04EE244 pKa = 4.16WQHH247 pKa = 6.67DD248 pKa = 3.83NLFQVTPYY256 pKa = 10.63TYY258 pKa = 10.09MLANHH263 pKa = 7.04HH264 pKa = 6.13CTSIEE269 pKa = 3.95EE270 pKa = 4.3ATSDD274 pKa = 3.97LQWMADD280 pKa = 3.28HH281 pKa = 7.05ARR283 pKa = 11.84TLRR286 pKa = 11.84SSTIGNRR293 pKa = 11.84ASTSLDD299 pKa = 3.21LQGQEE304 pKa = 4.01RR305 pKa = 11.84QPGPEE310 pKa = 3.6AA311 pKa = 4.32
MM1 pKa = 7.36RR2 pKa = 11.84TNANAVRR9 pKa = 11.84RR10 pKa = 11.84VEE12 pKa = 4.94FMTTPATTEE21 pKa = 4.01DD22 pKa = 3.79AFQSGTQEE30 pKa = 4.31SSDD33 pKa = 3.85SPSEE37 pKa = 4.44PIHH40 pKa = 5.16THH42 pKa = 3.12QTTWKK47 pKa = 9.41RR48 pKa = 11.84NFEE51 pKa = 4.06FRR53 pKa = 11.84SCNAFLTYY61 pKa = 9.53PRR63 pKa = 11.84CALDD67 pKa = 3.57CEE69 pKa = 4.63YY70 pKa = 11.46VLDD73 pKa = 4.73LPVGSYY79 pKa = 7.89VFRR82 pKa = 11.84WSPIYY87 pKa = 10.88ALVSAEE93 pKa = 3.76NHH95 pKa = 5.26QDD97 pKa = 3.26GGIHH101 pKa = 6.03LHH103 pKa = 6.7CLIQTLGRR111 pKa = 11.84ITTRR115 pKa = 11.84DD116 pKa = 3.27PAFFDD121 pKa = 5.1LDD123 pKa = 3.73NQYY126 pKa = 10.86HH127 pKa = 6.7PNIQAARR134 pKa = 11.84SPDD137 pKa = 3.22NTRR140 pKa = 11.84DD141 pKa = 3.81YY142 pKa = 11.06ILKK145 pKa = 10.47DD146 pKa = 3.85PISHH150 pKa = 5.64VEE152 pKa = 3.81RR153 pKa = 11.84GTFQPRR159 pKa = 11.84RR160 pKa = 11.84GNTRR164 pKa = 11.84NPGDD168 pKa = 3.75RR169 pKa = 11.84GSGSKK174 pKa = 10.26DD175 pKa = 3.09AIMKK179 pKa = 9.31EE180 pKa = 3.93IFRR183 pKa = 11.84TATSKK188 pKa = 10.91EE189 pKa = 3.99DD190 pKa = 3.83YY191 pKa = 10.93LRR193 pKa = 11.84MIQKK197 pKa = 9.61SFPFEE202 pKa = 3.95WAKK205 pKa = 10.73HH206 pKa = 4.91LSQFEE211 pKa = 3.98YY212 pKa = 10.21SANYY216 pKa = 9.33LFPPVPEE223 pKa = 4.46PFNNDD228 pKa = 3.09FPEE231 pKa = 4.38TEE233 pKa = 4.15PDD235 pKa = 4.28LICRR239 pKa = 11.84EE240 pKa = 4.27VLDD243 pKa = 4.04EE244 pKa = 4.16WQHH247 pKa = 6.67DD248 pKa = 3.83NLFQVTPYY256 pKa = 10.63TYY258 pKa = 10.09MLANHH263 pKa = 7.04HH264 pKa = 6.13CTSIEE269 pKa = 3.95EE270 pKa = 4.3ATSDD274 pKa = 3.97LQWMADD280 pKa = 3.28HH281 pKa = 7.05ARR283 pKa = 11.84TLRR286 pKa = 11.84SSTIGNRR293 pKa = 11.84ASTSLDD299 pKa = 3.21LQGQEE304 pKa = 4.01RR305 pKa = 11.84QPGPEE310 pKa = 3.6AA311 pKa = 4.32
Molecular weight: 35.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A218KH31|A0A218KH31_9GEMI Replication-associated protein OS=Sugarcane striate virus OX=1868659 PE=3 SV=1
MM1 pKa = 7.61SSRR4 pKa = 11.84GPSRR8 pKa = 11.84KK9 pKa = 9.25RR10 pKa = 11.84PRR12 pKa = 11.84SSTRR16 pKa = 11.84VVKK19 pKa = 10.11WPSEE23 pKa = 3.88ALRR26 pKa = 11.84KK27 pKa = 10.25GYY29 pKa = 8.32TPYY32 pKa = 11.08NSYY35 pKa = 11.18LRR37 pKa = 11.84GTKK40 pKa = 9.64SGRR43 pKa = 11.84SDD45 pKa = 3.22KK46 pKa = 10.99RR47 pKa = 11.84PNLQVQTFYY56 pKa = 11.63ACGPTQVQVKK66 pKa = 10.19KK67 pKa = 10.9GGIVNLLNGYY77 pKa = 7.14TRR79 pKa = 11.84GSDD82 pKa = 3.09DD83 pKa = 3.9NQRR86 pKa = 11.84HH87 pKa = 4.27TCEE90 pKa = 3.98TITYY94 pKa = 9.46KK95 pKa = 10.93LSLDD99 pKa = 3.36LHH101 pKa = 7.78INIATDD107 pKa = 3.86LFKK110 pKa = 11.35NCARR114 pKa = 11.84GTGVIWLIYY123 pKa = 8.55DD124 pKa = 3.99TSPTGTQLTTQQIFAYY140 pKa = 9.16PDD142 pKa = 3.47ALSLYY147 pKa = 8.8PYY149 pKa = 7.71TWKK152 pKa = 10.51VARR155 pKa = 11.84DD156 pKa = 3.32QCHH159 pKa = 5.62RR160 pKa = 11.84FIIKK164 pKa = 10.15RR165 pKa = 11.84RR166 pKa = 11.84WTFRR170 pKa = 11.84LEE172 pKa = 4.29SNGTLGSSDD181 pKa = 3.89YY182 pKa = 10.54SSKK185 pKa = 10.84PIAPCNNNVYY195 pKa = 10.07FSKK198 pKa = 10.23FVKK201 pKa = 10.3RR202 pKa = 11.84LGVRR206 pKa = 11.84TEE208 pKa = 3.85WKK210 pKa = 8.43NTTGGDD216 pKa = 3.48AGDD219 pKa = 3.88MKK221 pKa = 11.13SGGLFLAFAAGNGMLFNVDD240 pKa = 3.02GTTRR244 pKa = 11.84LYY246 pKa = 10.78FKK248 pKa = 10.41STGNQQ253 pKa = 2.93
MM1 pKa = 7.61SSRR4 pKa = 11.84GPSRR8 pKa = 11.84KK9 pKa = 9.25RR10 pKa = 11.84PRR12 pKa = 11.84SSTRR16 pKa = 11.84VVKK19 pKa = 10.11WPSEE23 pKa = 3.88ALRR26 pKa = 11.84KK27 pKa = 10.25GYY29 pKa = 8.32TPYY32 pKa = 11.08NSYY35 pKa = 11.18LRR37 pKa = 11.84GTKK40 pKa = 9.64SGRR43 pKa = 11.84SDD45 pKa = 3.22KK46 pKa = 10.99RR47 pKa = 11.84PNLQVQTFYY56 pKa = 11.63ACGPTQVQVKK66 pKa = 10.19KK67 pKa = 10.9GGIVNLLNGYY77 pKa = 7.14TRR79 pKa = 11.84GSDD82 pKa = 3.09DD83 pKa = 3.9NQRR86 pKa = 11.84HH87 pKa = 4.27TCEE90 pKa = 3.98TITYY94 pKa = 9.46KK95 pKa = 10.93LSLDD99 pKa = 3.36LHH101 pKa = 7.78INIATDD107 pKa = 3.86LFKK110 pKa = 11.35NCARR114 pKa = 11.84GTGVIWLIYY123 pKa = 8.55DD124 pKa = 3.99TSPTGTQLTTQQIFAYY140 pKa = 9.16PDD142 pKa = 3.47ALSLYY147 pKa = 8.8PYY149 pKa = 7.71TWKK152 pKa = 10.51VARR155 pKa = 11.84DD156 pKa = 3.32QCHH159 pKa = 5.62RR160 pKa = 11.84FIIKK164 pKa = 10.15RR165 pKa = 11.84RR166 pKa = 11.84WTFRR170 pKa = 11.84LEE172 pKa = 4.29SNGTLGSSDD181 pKa = 3.89YY182 pKa = 10.54SSKK185 pKa = 10.84PIAPCNNNVYY195 pKa = 10.07FSKK198 pKa = 10.23FVKK201 pKa = 10.3RR202 pKa = 11.84LGVRR206 pKa = 11.84TEE208 pKa = 3.85WKK210 pKa = 8.43NTTGGDD216 pKa = 3.48AGDD219 pKa = 3.88MKK221 pKa = 11.13SGGLFLAFAAGNGMLFNVDD240 pKa = 3.02GTTRR244 pKa = 11.84LYY246 pKa = 10.78FKK248 pKa = 10.41STGNQQ253 pKa = 2.93
Molecular weight: 28.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1048 |
96 |
388 |
262.0 |
29.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.534 ± 0.424 | 2.29 ± 0.237 |
6.107 ± 0.943 | 4.676 ± 1.116 |
5.63 ± 0.532 | 6.202 ± 1.573 |
2.576 ± 0.604 | 4.962 ± 0.415 |
4.103 ± 0.901 | 6.775 ± 0.738 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.527 ± 0.17 | 5.153 ± 0.669 |
6.679 ± 0.904 | 4.771 ± 0.31 |
7.156 ± 0.284 | 7.156 ± 0.411 |
8.206 ± 0.75 | 4.389 ± 0.717 |
1.908 ± 0.103 | 4.198 ± 0.455 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
