
Gokushovirus WZ-2015a
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
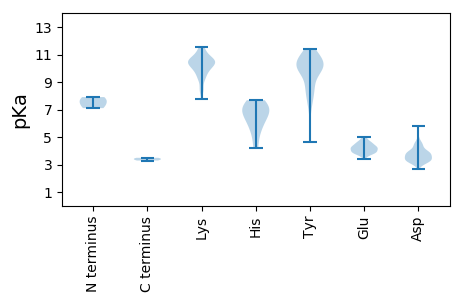
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
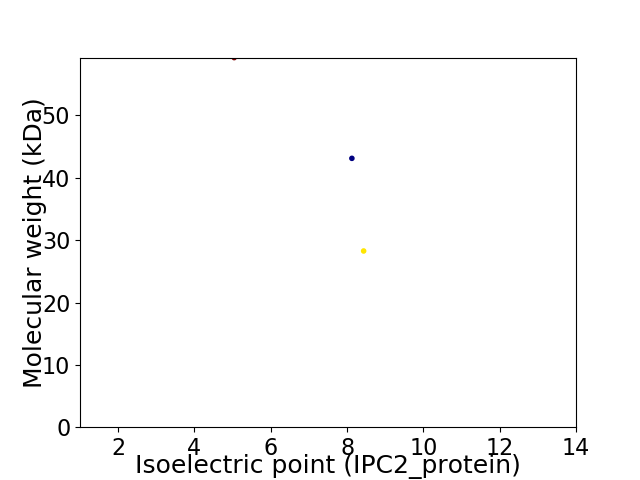
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L2BLB6|A0A1L2BLB6_9VIRU VP2 OS=Gokushovirus WZ-2015a OX=1758150 PE=4 SV=1
MM1 pKa = 7.91AYY3 pKa = 9.84QMKK6 pKa = 9.58MPSSVVNNSNILPQATVNLSTFDD29 pKa = 3.67LSEE32 pKa = 4.13TLVTGIGTDD41 pKa = 3.22NLYY44 pKa = 10.06PVYY47 pKa = 9.92WRR49 pKa = 11.84EE50 pKa = 3.99LQPNDD55 pKa = 3.15HH56 pKa = 6.82FEE58 pKa = 4.35IKK60 pKa = 10.48VSALSRR66 pKa = 11.84IMPMVAPPMTNIKK79 pKa = 10.56LKK81 pKa = 10.67FFAFWVPNRR90 pKa = 11.84TIWSHH95 pKa = 4.17WMNFMGEE102 pKa = 3.94EE103 pKa = 4.68TYY105 pKa = 10.84QGDD108 pKa = 3.93KK109 pKa = 10.92VDD111 pKa = 3.59VRR113 pKa = 11.84VPQIKK118 pKa = 9.23MKK120 pKa = 10.6SAANVTNGIADD131 pKa = 3.87YY132 pKa = 11.05LGIPPTVDD140 pKa = 3.22SIDD143 pKa = 3.76YY144 pKa = 8.29TVSALPFRR152 pKa = 11.84AYY154 pKa = 10.9NKK156 pKa = 9.46IWNEE160 pKa = 3.69WFRR163 pKa = 11.84ANYY166 pKa = 8.72IQDD169 pKa = 3.54VVQEE173 pKa = 4.27FSGDD177 pKa = 3.45TSDD180 pKa = 5.81EE181 pKa = 4.17EE182 pKa = 4.41TLDD185 pKa = 3.97KK186 pKa = 11.45YY187 pKa = 11.37KK188 pKa = 10.66LLKK191 pKa = 9.74KK192 pKa = 10.28GKK194 pKa = 8.65PIDD197 pKa = 3.94YY198 pKa = 7.46FTSCLPQPEE207 pKa = 4.18QVDD210 pKa = 3.31ISLLGNAPVKK220 pKa = 8.54TTTLPVKK227 pKa = 10.57LNYY230 pKa = 9.1QASGLNYY237 pKa = 10.19RR238 pKa = 11.84GTPYY242 pKa = 10.63TDD244 pKa = 3.35GGSDD248 pKa = 3.55LQISNDD254 pKa = 3.43GGSFWGTDD262 pKa = 3.02TNMFADD268 pKa = 4.38MSAVSGLSIEE278 pKa = 4.57SLRR281 pKa = 11.84KK282 pKa = 9.36ASSIQVLLEE291 pKa = 3.41KK292 pKa = 10.53DD293 pKa = 3.17RR294 pKa = 11.84RR295 pKa = 11.84SGLRR299 pKa = 11.84YY300 pKa = 8.45IDD302 pKa = 4.7LIKK305 pKa = 10.59EE306 pKa = 4.15HH307 pKa = 7.09FGMEE311 pKa = 3.87IPDD314 pKa = 3.8FLVGRR319 pKa = 11.84SEE321 pKa = 4.38YY322 pKa = 10.29VGSVTIPINSEE333 pKa = 4.05PIVQQSEE340 pKa = 4.63SASTPQGNLASLGFGVGVDD359 pKa = 3.67KK360 pKa = 11.22LCEE363 pKa = 3.94FSARR367 pKa = 11.84EE368 pKa = 4.07HH369 pKa = 5.22GHH371 pKa = 7.68LMILACVEE379 pKa = 4.23SEE381 pKa = 4.51VIYY384 pKa = 10.59QQGLDD389 pKa = 3.8RR390 pKa = 11.84KK391 pKa = 8.29WEE393 pKa = 3.82KK394 pKa = 10.38RR395 pKa = 11.84DD396 pKa = 3.3RR397 pKa = 11.84FDD399 pKa = 3.85YY400 pKa = 10.53YY401 pKa = 11.03FPEE404 pKa = 4.39FANLGEE410 pKa = 4.14QPVMEE415 pKa = 4.97RR416 pKa = 11.84EE417 pKa = 4.19LMLTASNATTDD428 pKa = 3.29KK429 pKa = 11.11VFGYY433 pKa = 7.33TPRR436 pKa = 11.84YY437 pKa = 8.0RR438 pKa = 11.84EE439 pKa = 3.66EE440 pKa = 4.09RR441 pKa = 11.84EE442 pKa = 4.43GYY444 pKa = 10.31NKK446 pKa = 9.93ISGIMRR452 pKa = 11.84SNVTSGFQSLDD463 pKa = 3.18VWHH466 pKa = 7.19LAQKK470 pKa = 10.02FAAPPQLNGEE480 pKa = 4.6FIEE483 pKa = 4.65SNSPLEE489 pKa = 3.98RR490 pKa = 11.84VLAVSEE496 pKa = 4.92DD497 pKa = 3.31IFVLLNIAWDD507 pKa = 3.64IKK509 pKa = 9.45ATRR512 pKa = 11.84SIPIQSNPSLVTGKK526 pKa = 10.46LL527 pKa = 3.29
MM1 pKa = 7.91AYY3 pKa = 9.84QMKK6 pKa = 9.58MPSSVVNNSNILPQATVNLSTFDD29 pKa = 3.67LSEE32 pKa = 4.13TLVTGIGTDD41 pKa = 3.22NLYY44 pKa = 10.06PVYY47 pKa = 9.92WRR49 pKa = 11.84EE50 pKa = 3.99LQPNDD55 pKa = 3.15HH56 pKa = 6.82FEE58 pKa = 4.35IKK60 pKa = 10.48VSALSRR66 pKa = 11.84IMPMVAPPMTNIKK79 pKa = 10.56LKK81 pKa = 10.67FFAFWVPNRR90 pKa = 11.84TIWSHH95 pKa = 4.17WMNFMGEE102 pKa = 3.94EE103 pKa = 4.68TYY105 pKa = 10.84QGDD108 pKa = 3.93KK109 pKa = 10.92VDD111 pKa = 3.59VRR113 pKa = 11.84VPQIKK118 pKa = 9.23MKK120 pKa = 10.6SAANVTNGIADD131 pKa = 3.87YY132 pKa = 11.05LGIPPTVDD140 pKa = 3.22SIDD143 pKa = 3.76YY144 pKa = 8.29TVSALPFRR152 pKa = 11.84AYY154 pKa = 10.9NKK156 pKa = 9.46IWNEE160 pKa = 3.69WFRR163 pKa = 11.84ANYY166 pKa = 8.72IQDD169 pKa = 3.54VVQEE173 pKa = 4.27FSGDD177 pKa = 3.45TSDD180 pKa = 5.81EE181 pKa = 4.17EE182 pKa = 4.41TLDD185 pKa = 3.97KK186 pKa = 11.45YY187 pKa = 11.37KK188 pKa = 10.66LLKK191 pKa = 9.74KK192 pKa = 10.28GKK194 pKa = 8.65PIDD197 pKa = 3.94YY198 pKa = 7.46FTSCLPQPEE207 pKa = 4.18QVDD210 pKa = 3.31ISLLGNAPVKK220 pKa = 8.54TTTLPVKK227 pKa = 10.57LNYY230 pKa = 9.1QASGLNYY237 pKa = 10.19RR238 pKa = 11.84GTPYY242 pKa = 10.63TDD244 pKa = 3.35GGSDD248 pKa = 3.55LQISNDD254 pKa = 3.43GGSFWGTDD262 pKa = 3.02TNMFADD268 pKa = 4.38MSAVSGLSIEE278 pKa = 4.57SLRR281 pKa = 11.84KK282 pKa = 9.36ASSIQVLLEE291 pKa = 3.41KK292 pKa = 10.53DD293 pKa = 3.17RR294 pKa = 11.84RR295 pKa = 11.84SGLRR299 pKa = 11.84YY300 pKa = 8.45IDD302 pKa = 4.7LIKK305 pKa = 10.59EE306 pKa = 4.15HH307 pKa = 7.09FGMEE311 pKa = 3.87IPDD314 pKa = 3.8FLVGRR319 pKa = 11.84SEE321 pKa = 4.38YY322 pKa = 10.29VGSVTIPINSEE333 pKa = 4.05PIVQQSEE340 pKa = 4.63SASTPQGNLASLGFGVGVDD359 pKa = 3.67KK360 pKa = 11.22LCEE363 pKa = 3.94FSARR367 pKa = 11.84EE368 pKa = 4.07HH369 pKa = 5.22GHH371 pKa = 7.68LMILACVEE379 pKa = 4.23SEE381 pKa = 4.51VIYY384 pKa = 10.59QQGLDD389 pKa = 3.8RR390 pKa = 11.84KK391 pKa = 8.29WEE393 pKa = 3.82KK394 pKa = 10.38RR395 pKa = 11.84DD396 pKa = 3.3RR397 pKa = 11.84FDD399 pKa = 3.85YY400 pKa = 10.53YY401 pKa = 11.03FPEE404 pKa = 4.39FANLGEE410 pKa = 4.14QPVMEE415 pKa = 4.97RR416 pKa = 11.84EE417 pKa = 4.19LMLTASNATTDD428 pKa = 3.29KK429 pKa = 11.11VFGYY433 pKa = 7.33TPRR436 pKa = 11.84YY437 pKa = 8.0RR438 pKa = 11.84EE439 pKa = 3.66EE440 pKa = 4.09RR441 pKa = 11.84EE442 pKa = 4.43GYY444 pKa = 10.31NKK446 pKa = 9.93ISGIMRR452 pKa = 11.84SNVTSGFQSLDD463 pKa = 3.18VWHH466 pKa = 7.19LAQKK470 pKa = 10.02FAAPPQLNGEE480 pKa = 4.6FIEE483 pKa = 4.65SNSPLEE489 pKa = 3.98RR490 pKa = 11.84VLAVSEE496 pKa = 4.92DD497 pKa = 3.31IFVLLNIAWDD507 pKa = 3.64IKK509 pKa = 9.45ATRR512 pKa = 11.84SIPIQSNPSLVTGKK526 pKa = 10.46LL527 pKa = 3.29
Molecular weight: 59.28 kDa
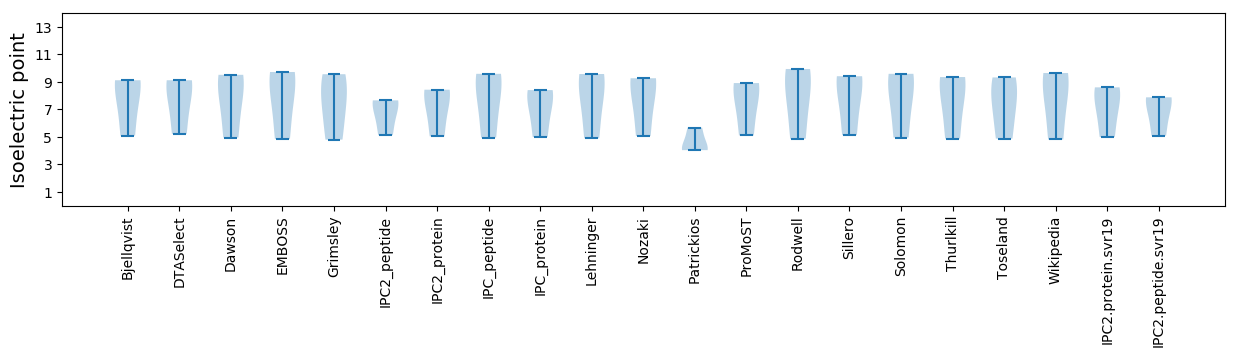
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L2BLB6|A0A1L2BLB6_9VIRU VP2 OS=Gokushovirus WZ-2015a OX=1758150 PE=4 SV=1
MM1 pKa = 7.54GFLSGLGSAVGGVVSSVAGGLFNKK25 pKa = 10.35NEE27 pKa = 3.96ADD29 pKa = 4.39KK30 pKa = 11.48NRR32 pKa = 11.84GWQEE36 pKa = 3.78DD37 pKa = 3.69MSNTSVQRR45 pKa = 11.84RR46 pKa = 11.84VADD49 pKa = 4.48LKK51 pKa = 11.3AAGLNPLLAVSNASSGASTPSGAQASANFDD81 pKa = 3.53SAGQAISTAIQLGMQRR97 pKa = 11.84DD98 pKa = 3.96VNNANINKK106 pKa = 8.49TNAEE110 pKa = 3.87AMKK113 pKa = 10.24IQDD116 pKa = 4.46DD117 pKa = 3.82IKK119 pKa = 9.73TNEE122 pKa = 3.8INRR125 pKa = 11.84LNTQMDD131 pKa = 3.96TKK133 pKa = 10.42LKK135 pKa = 9.77EE136 pKa = 3.9MGVKK140 pKa = 9.96SEE142 pKa = 4.57EE143 pKa = 4.06IKK145 pKa = 10.62QQLLAAQTDD154 pKa = 3.87QARR157 pKa = 11.84SNVLKK162 pKa = 9.95TIRR165 pKa = 11.84EE166 pKa = 4.25TKK168 pKa = 10.41NLSTQEE174 pKa = 3.91KK175 pKa = 10.65SMLIDD180 pKa = 3.43ISRR183 pKa = 11.84QIWEE187 pKa = 4.33LEE189 pKa = 3.78RR190 pKa = 11.84DD191 pKa = 3.71KK192 pKa = 11.52KK193 pKa = 11.08DD194 pKa = 3.17IANTEE199 pKa = 3.5SGMRR203 pKa = 11.84DD204 pKa = 2.67KK205 pKa = 10.77HH206 pKa = 5.8YY207 pKa = 11.15RR208 pKa = 11.84NVSPTNAYY216 pKa = 8.55QAAYY220 pKa = 7.82MAGRR224 pKa = 11.84KK225 pKa = 9.83GYY227 pKa = 10.57DD228 pKa = 3.9LIDD231 pKa = 3.77EE232 pKa = 4.84KK233 pKa = 11.5LNSDD237 pKa = 4.01SGFSKK242 pKa = 9.17WWSNFKK248 pKa = 10.44RR249 pKa = 11.84YY250 pKa = 9.74NKK252 pKa = 10.22EE253 pKa = 3.75SFPQQ257 pKa = 3.39
MM1 pKa = 7.54GFLSGLGSAVGGVVSSVAGGLFNKK25 pKa = 10.35NEE27 pKa = 3.96ADD29 pKa = 4.39KK30 pKa = 11.48NRR32 pKa = 11.84GWQEE36 pKa = 3.78DD37 pKa = 3.69MSNTSVQRR45 pKa = 11.84RR46 pKa = 11.84VADD49 pKa = 4.48LKK51 pKa = 11.3AAGLNPLLAVSNASSGASTPSGAQASANFDD81 pKa = 3.53SAGQAISTAIQLGMQRR97 pKa = 11.84DD98 pKa = 3.96VNNANINKK106 pKa = 8.49TNAEE110 pKa = 3.87AMKK113 pKa = 10.24IQDD116 pKa = 4.46DD117 pKa = 3.82IKK119 pKa = 9.73TNEE122 pKa = 3.8INRR125 pKa = 11.84LNTQMDD131 pKa = 3.96TKK133 pKa = 10.42LKK135 pKa = 9.77EE136 pKa = 3.9MGVKK140 pKa = 9.96SEE142 pKa = 4.57EE143 pKa = 4.06IKK145 pKa = 10.62QQLLAAQTDD154 pKa = 3.87QARR157 pKa = 11.84SNVLKK162 pKa = 9.95TIRR165 pKa = 11.84EE166 pKa = 4.25TKK168 pKa = 10.41NLSTQEE174 pKa = 3.91KK175 pKa = 10.65SMLIDD180 pKa = 3.43ISRR183 pKa = 11.84QIWEE187 pKa = 4.33LEE189 pKa = 3.78RR190 pKa = 11.84DD191 pKa = 3.71KK192 pKa = 11.52KK193 pKa = 11.08DD194 pKa = 3.17IANTEE199 pKa = 3.5SGMRR203 pKa = 11.84DD204 pKa = 2.67KK205 pKa = 10.77HH206 pKa = 5.8YY207 pKa = 11.15RR208 pKa = 11.84NVSPTNAYY216 pKa = 8.55QAAYY220 pKa = 7.82MAGRR224 pKa = 11.84KK225 pKa = 9.83GYY227 pKa = 10.57DD228 pKa = 3.9LIDD231 pKa = 3.77EE232 pKa = 4.84KK233 pKa = 11.5LNSDD237 pKa = 4.01SGFSKK242 pKa = 9.17WWSNFKK248 pKa = 10.44RR249 pKa = 11.84YY250 pKa = 9.74NKK252 pKa = 10.22EE253 pKa = 3.75SFPQQ257 pKa = 3.39
Molecular weight: 28.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1143 |
257 |
527 |
381.0 |
43.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.862 ± 1.464 | 1.312 ± 0.813 |
5.687 ± 0.197 | 7.087 ± 0.958 |
3.85 ± 0.57 | 6.037 ± 0.526 |
1.05 ± 0.223 | 5.512 ± 0.467 |
8.049 ± 2.083 | 7.612 ± 0.577 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.887 ± 0.277 | 6.212 ± 0.919 |
3.937 ± 1.163 | 4.374 ± 0.63 |
4.899 ± 0.343 | 7.612 ± 1.386 |
5.162 ± 0.345 | 6.037 ± 0.719 |
1.837 ± 0.094 | 4.987 ± 1.499 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
