
Rathayibacter tanaceti
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Rathayibacter
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
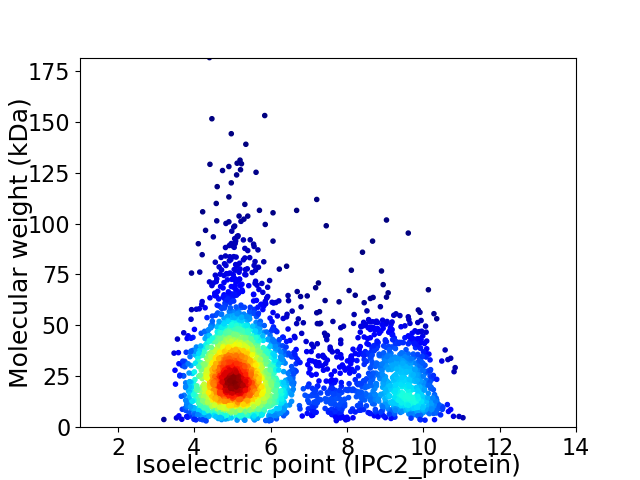
Virtual 2D-PAGE plot for 3209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A162G0P6|A0A162G0P6_9MICO 50S ribosomal protein L31 type B OS=Rathayibacter tanaceti OX=1671680 GN=rpmE2 PE=3 SV=1
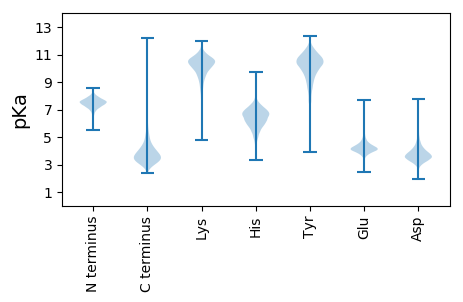
MM1 pKa = 7.62NITRR5 pKa = 11.84TARR8 pKa = 11.84WSAALVATGMLVTLTACSSDD28 pKa = 3.35SSSDD32 pKa = 3.61GASGSSDD39 pKa = 2.9GLTVLIASSSDD50 pKa = 3.32AEE52 pKa = 4.51GAAVTAANDD61 pKa = 2.87AWATEE66 pKa = 4.0NGTTVDD72 pKa = 3.56TVIASDD78 pKa = 3.95LTQQLGQGFSGDD90 pKa = 3.61NPPDD94 pKa = 3.81LFYY97 pKa = 10.88MSWDD101 pKa = 3.3QFQTYY106 pKa = 10.42ASNRR110 pKa = 11.84YY111 pKa = 9.0LEE113 pKa = 5.43PYY115 pKa = 9.72AQDD118 pKa = 4.1AEE120 pKa = 4.42NADD123 pKa = 3.53AFYY126 pKa = 9.72PALKK130 pKa = 10.5DD131 pKa = 3.27AFSYY135 pKa = 11.2DD136 pKa = 3.36DD137 pKa = 4.01QFYY140 pKa = 11.1CEE142 pKa = 4.63PKK144 pKa = 10.69DD145 pKa = 3.91FSTLGLVINTDD156 pKa = 2.83LWAAAGLTDD165 pKa = 5.62ADD167 pKa = 5.36IPTDD171 pKa = 3.48WDD173 pKa = 3.82SLEE176 pKa = 4.2SAAQKK181 pKa = 9.34LTTDD185 pKa = 3.83TVAGLSFGPEE195 pKa = 3.8YY196 pKa = 11.14ARR198 pKa = 11.84IGTFMNQAGGSLISEE213 pKa = 5.23DD214 pKa = 3.57GTTATANTPEE224 pKa = 4.17NVAGLQEE231 pKa = 4.23VQKK234 pKa = 10.73LLSDD238 pKa = 3.92GVLRR242 pKa = 11.84FPADD246 pKa = 3.39IDD248 pKa = 4.26SGWSGEE254 pKa = 4.11AFGKK258 pKa = 10.22GAAAMVIEE266 pKa = 4.87GPWINGALKK275 pKa = 10.45ADD277 pKa = 3.97YY278 pKa = 10.22PDD280 pKa = 3.26VKK282 pKa = 10.31YY283 pKa = 9.29TIAEE287 pKa = 4.24LPAGPGGKK295 pKa = 7.98STFTFSNCWGIPAGSDD311 pKa = 2.99TAEE314 pKa = 4.18SAEE317 pKa = 4.26SLVSYY322 pKa = 7.74LTSDD326 pKa = 3.59EE327 pKa = 4.12QQLAFSDD334 pKa = 3.87AFGVIPSTEE343 pKa = 3.83TGAAEE348 pKa = 4.21YY349 pKa = 8.03ATKK352 pKa = 10.74YY353 pKa = 9.6PEE355 pKa = 3.84NAAFVAGNDD364 pKa = 3.81YY365 pKa = 11.05AVSPVAFAGAATVITDD381 pKa = 3.94FNASLEE387 pKa = 4.71GIATADD393 pKa = 3.42PEE395 pKa = 5.33AILEE399 pKa = 4.29TFQTNLQDD407 pKa = 5.44ALDD410 pKa = 4.02TANAKK415 pKa = 9.81
MM1 pKa = 7.62NITRR5 pKa = 11.84TARR8 pKa = 11.84WSAALVATGMLVTLTACSSDD28 pKa = 3.35SSSDD32 pKa = 3.61GASGSSDD39 pKa = 2.9GLTVLIASSSDD50 pKa = 3.32AEE52 pKa = 4.51GAAVTAANDD61 pKa = 2.87AWATEE66 pKa = 4.0NGTTVDD72 pKa = 3.56TVIASDD78 pKa = 3.95LTQQLGQGFSGDD90 pKa = 3.61NPPDD94 pKa = 3.81LFYY97 pKa = 10.88MSWDD101 pKa = 3.3QFQTYY106 pKa = 10.42ASNRR110 pKa = 11.84YY111 pKa = 9.0LEE113 pKa = 5.43PYY115 pKa = 9.72AQDD118 pKa = 4.1AEE120 pKa = 4.42NADD123 pKa = 3.53AFYY126 pKa = 9.72PALKK130 pKa = 10.5DD131 pKa = 3.27AFSYY135 pKa = 11.2DD136 pKa = 3.36DD137 pKa = 4.01QFYY140 pKa = 11.1CEE142 pKa = 4.63PKK144 pKa = 10.69DD145 pKa = 3.91FSTLGLVINTDD156 pKa = 2.83LWAAAGLTDD165 pKa = 5.62ADD167 pKa = 5.36IPTDD171 pKa = 3.48WDD173 pKa = 3.82SLEE176 pKa = 4.2SAAQKK181 pKa = 9.34LTTDD185 pKa = 3.83TVAGLSFGPEE195 pKa = 3.8YY196 pKa = 11.14ARR198 pKa = 11.84IGTFMNQAGGSLISEE213 pKa = 5.23DD214 pKa = 3.57GTTATANTPEE224 pKa = 4.17NVAGLQEE231 pKa = 4.23VQKK234 pKa = 10.73LLSDD238 pKa = 3.92GVLRR242 pKa = 11.84FPADD246 pKa = 3.39IDD248 pKa = 4.26SGWSGEE254 pKa = 4.11AFGKK258 pKa = 10.22GAAAMVIEE266 pKa = 4.87GPWINGALKK275 pKa = 10.45ADD277 pKa = 3.97YY278 pKa = 10.22PDD280 pKa = 3.26VKK282 pKa = 10.31YY283 pKa = 9.29TIAEE287 pKa = 4.24LPAGPGGKK295 pKa = 7.98STFTFSNCWGIPAGSDD311 pKa = 2.99TAEE314 pKa = 4.18SAEE317 pKa = 4.26SLVSYY322 pKa = 7.74LTSDD326 pKa = 3.59EE327 pKa = 4.12QQLAFSDD334 pKa = 3.87AFGVIPSTEE343 pKa = 3.83TGAAEE348 pKa = 4.21YY349 pKa = 8.03ATKK352 pKa = 10.74YY353 pKa = 9.6PEE355 pKa = 3.84NAAFVAGNDD364 pKa = 3.81YY365 pKa = 11.05AVSPVAFAGAATVITDD381 pKa = 3.94FNASLEE387 pKa = 4.71GIATADD393 pKa = 3.42PEE395 pKa = 5.33AILEE399 pKa = 4.29TFQTNLQDD407 pKa = 5.44ALDD410 pKa = 4.02TANAKK415 pKa = 9.81
Molecular weight: 43.27 kDa
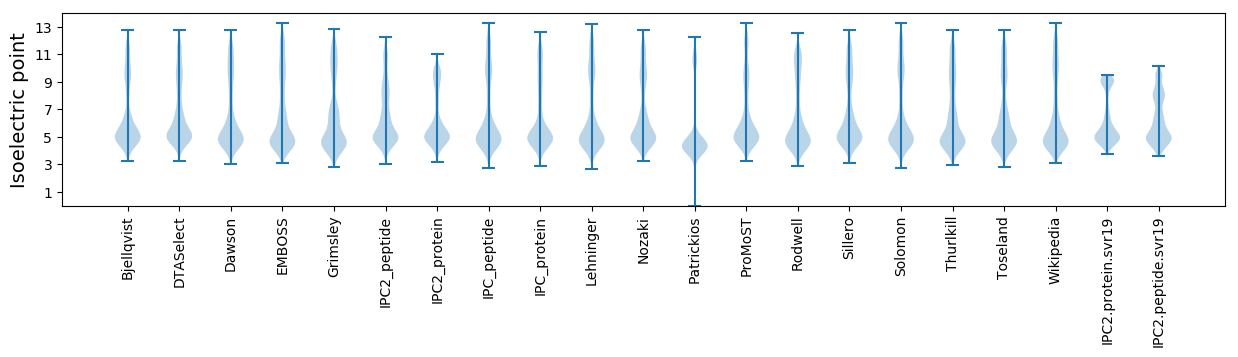
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166HZK1|A0A166HZK1_9MICO L-lysine cyclodeaminase OS=Rathayibacter tanaceti OX=1671680 GN=rapL PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.73SMKK11 pKa = 9.64SQPGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 8.59LNPRR34 pKa = 11.84FKK36 pKa = 10.79ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.73SMKK11 pKa = 9.64SQPGSQVVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 8.59LNPRR34 pKa = 11.84FKK36 pKa = 10.79ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
878195 |
29 |
1757 |
273.7 |
29.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.304 ± 0.065 | 0.563 ± 0.012 |
5.918 ± 0.039 | 5.848 ± 0.051 |
2.926 ± 0.029 | 9.111 ± 0.044 |
1.938 ± 0.022 | 4.045 ± 0.031 |
1.679 ± 0.032 | 10.36 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.586 ± 0.016 | 1.736 ± 0.024 |
5.524 ± 0.033 | 2.589 ± 0.022 |
8.026 ± 0.063 | 6.394 ± 0.043 |
6.073 ± 0.05 | 9.126 ± 0.044 |
1.379 ± 0.021 | 1.878 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
