
Pseudidiomarina atlantica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Pseudidiomarina
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
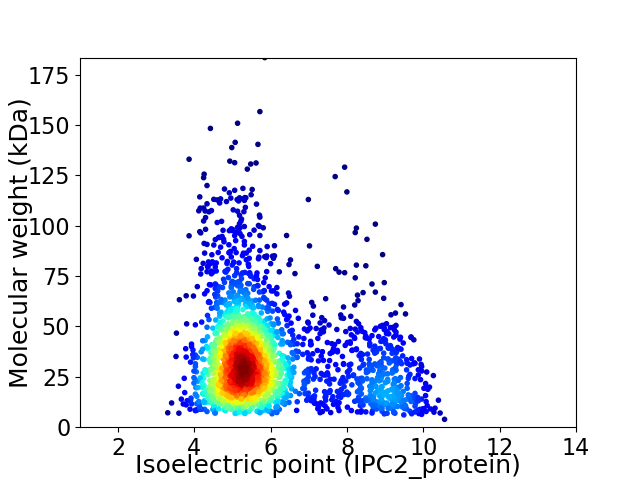
Virtual 2D-PAGE plot for 2290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A094J8Z5|A0A094J8Z5_9GAMM LysR family transcriptional regulator OS=Pseudidiomarina atlantica OX=1517416 GN=IDAT_05205 PE=3 SV=1
MM1 pKa = 7.21VSNNKK6 pKa = 9.71FIYY9 pKa = 10.19GLTFALILLSGSAQAATYY27 pKa = 10.23DD28 pKa = 3.63FRR30 pKa = 11.84KK31 pKa = 10.01NLPPGCSQKK40 pKa = 10.96GNSPAQCPNGLTLQWNDD57 pKa = 2.91VLVFDD62 pKa = 6.1KK63 pKa = 10.74IAEE66 pKa = 4.27VNVTGNVWLSNAQVFLGANTPLTINATGSISSDD99 pKa = 2.41STMQMFGDD107 pKa = 4.6LNAVGQISLGSDD119 pKa = 3.02NQVQGNLNSQNQITLNNNTDD139 pKa = 3.33VTGSLVAPQVSLVSSNTITGPISATQLTLGFNSQVTGNLNVTNNLSVGSSSSITGSINAGNLQSNSPVSLNGPIVVDD216 pKa = 3.54NEE218 pKa = 3.93FNLASGSTVNGSVSANRR235 pKa = 11.84VTLAPSNSTLTGDD248 pKa = 3.28VTANEE253 pKa = 3.97NVIIGSGNTIDD264 pKa = 4.24GNVAAGFIRR273 pKa = 11.84LEE275 pKa = 4.25ASNATITGNAIATGNIVIDD294 pKa = 3.41WAGQIGGDD302 pKa = 3.34ATAVNITNNAGNTDD316 pKa = 3.91SVGGTAFCDD325 pKa = 3.7TSDD328 pKa = 3.44GAQPLSCQTGSGGGGATQCDD348 pKa = 4.14ALNDD352 pKa = 3.87LVDD355 pKa = 3.61YY356 pKa = 10.85GVIGDD361 pKa = 4.09SGFTYY366 pKa = 10.81GNNSEE371 pKa = 4.29INGTEE376 pKa = 3.44IDD378 pKa = 5.1DD379 pKa = 4.58PNNTNGNTPTPTGVVEE395 pKa = 4.11EE396 pKa = 5.0AVVQFPPLTPNVFPSFNGGNSFTNPANLGPGTYY429 pKa = 10.38NEE431 pKa = 4.84INLSGNNASASLTGGGTYY449 pKa = 10.3QIDD452 pKa = 4.19SINFGPQSQTLTLGPGDD469 pKa = 3.61YY470 pKa = 10.26FVRR473 pKa = 11.84TISLNNDD480 pKa = 2.95STIAIAPAGPVRR492 pKa = 11.84IFIEE496 pKa = 4.51DD497 pKa = 4.07SITGGNNLFFNPTGAVQNLMINLYY521 pKa = 10.36DD522 pKa = 3.82GASFEE527 pKa = 4.46IGNYY531 pKa = 8.27NQGSTTYY538 pKa = 9.24TFNGVLYY545 pKa = 10.64APYY548 pKa = 9.43TSNDD552 pKa = 2.55ITFGNNSNLQGAVLSAGTVDD572 pKa = 3.09IGNNTSINFTDD583 pKa = 3.78AVQDD587 pKa = 3.66AVRR590 pKa = 11.84DD591 pKa = 3.78AFGCDD596 pKa = 3.1TEE598 pKa = 4.56VTDD601 pKa = 3.15IHH603 pKa = 7.0HH604 pKa = 6.46YY605 pKa = 10.21RR606 pKa = 11.84IQHH609 pKa = 5.81PLSVVSCVAAPVQVVACLDD628 pKa = 3.47ASCASRR634 pKa = 11.84YY635 pKa = 8.26TDD637 pKa = 3.15ATEE640 pKa = 3.99VALSSSATGSTFANGGVLGFTGGQGTLGLRR670 pKa = 11.84YY671 pKa = 9.72VDD673 pKa = 4.48GGTTTVSLSNGAPAAQNPTEE693 pKa = 4.9CYY695 pKa = 10.35DD696 pKa = 3.65SAGTATTNCEE706 pKa = 3.66IEE708 pKa = 4.2FRR710 pKa = 11.84TAGLIFTANDD720 pKa = 3.76GLSAIPPSFAGTDD733 pKa = 3.8FDD735 pKa = 4.01VLARR739 pKa = 11.84AVEE742 pKa = 4.57TNTQTGACEE751 pKa = 3.77ARR753 pKa = 11.84LTGAQVLEE761 pKa = 4.54LATSCRR767 pKa = 11.84NPLTCQAGQNFIANGANVPLNDD789 pKa = 3.8AGATLSYY796 pKa = 11.31ANVPVTFDD804 pKa = 3.58ANGNAPLVANYY815 pKa = 9.68TDD817 pKa = 3.55VGALRR822 pKa = 11.84LHH824 pKa = 6.09GRR826 pKa = 11.84VNLSEE831 pKa = 4.68APNADD836 pKa = 3.35EE837 pKa = 6.05SVIDD841 pKa = 4.75DD842 pKa = 4.09PAITLTGTSVNDD854 pKa = 3.73FVVKK858 pKa = 9.9PHH860 pKa = 5.69TLRR863 pKa = 11.84LFAVDD868 pKa = 3.78NANQAVSATTSGGSAFATAGDD889 pKa = 4.08NFSLIITAEE898 pKa = 4.04NANGDD903 pKa = 3.48ATPNFGRR910 pKa = 11.84EE911 pKa = 3.82TPAAEE916 pKa = 4.06AQASYY921 pKa = 11.11VQTLYY926 pKa = 10.49PSTATAPASDD936 pKa = 3.88FVGTQGWQLDD946 pKa = 3.94TNRR949 pKa = 11.84AGSLRR954 pKa = 11.84TDD956 pKa = 3.23SARR959 pKa = 11.84WLDD962 pKa = 3.4VGTIEE967 pKa = 5.65LAPGLNGNSYY977 pKa = 10.81LGAGDD982 pKa = 3.63VAAKK986 pKa = 10.36QNATVGRR993 pKa = 11.84FAPFRR998 pKa = 11.84FALNTSAVANSCVAGDD1014 pKa = 3.95FTYY1017 pKa = 10.2MSEE1020 pKa = 4.04PALTVSYY1027 pKa = 10.42EE1028 pKa = 4.03LYY1030 pKa = 10.94AVDD1033 pKa = 3.38SDD1035 pKa = 4.58GNITEE1040 pKa = 5.03NYY1042 pKa = 9.57DD1043 pKa = 3.17SSGYY1047 pKa = 10.59SGTAEE1052 pKa = 3.94VGFAAANLAPADD1064 pKa = 3.76TATDD1068 pKa = 3.74RR1069 pKa = 11.84FADD1072 pKa = 3.84RR1073 pKa = 11.84LLNLAAQQAWTAGVMTFNAANAGFNRR1099 pKa = 11.84HH1100 pKa = 6.21PDD1102 pKa = 3.48GNIDD1106 pKa = 3.81GPFASLQLGLRR1117 pKa = 11.84VLSEE1121 pKa = 3.4IDD1123 pKa = 3.24NRR1125 pKa = 11.84NFAGSDD1131 pKa = 3.23LTLAAVAGAAAPLNGTAQLRR1151 pKa = 11.84YY1152 pKa = 9.96GRR1154 pKa = 11.84LVLEE1158 pKa = 4.12NTYY1161 pKa = 10.78GPEE1164 pKa = 3.99NEE1166 pKa = 4.67DD1167 pKa = 3.74LSVPLYY1173 pKa = 10.99AEE1175 pKa = 4.18YY1176 pKa = 10.6FDD1178 pKa = 3.91GSRR1181 pKa = 11.84FVTHH1185 pKa = 7.11TDD1187 pKa = 3.6DD1188 pKa = 3.83SCSSALVAGLNVIADD1203 pKa = 4.36PNNLTPVAAGSDD1215 pKa = 3.62STLTDD1220 pKa = 3.17GALPFDD1226 pKa = 4.15TLRR1229 pKa = 11.84WLAPGTGNTGEE1240 pKa = 4.72FIYY1243 pKa = 10.41EE1244 pKa = 3.69YY1245 pKa = 10.49DD1246 pKa = 3.41APAWLEE1252 pKa = 3.93FEE1254 pKa = 4.37WRR1256 pKa = 11.84DD1257 pKa = 3.43EE1258 pKa = 4.15AGAMYY1263 pKa = 10.56LDD1265 pKa = 4.02PRR1267 pKa = 11.84AIGGFGQYY1275 pKa = 9.73RR1276 pKa = 11.84GNSRR1280 pKa = 11.84VLFWKK1285 pKa = 10.07EE1286 pKa = 3.54VNN1288 pKa = 3.47
MM1 pKa = 7.21VSNNKK6 pKa = 9.71FIYY9 pKa = 10.19GLTFALILLSGSAQAATYY27 pKa = 10.23DD28 pKa = 3.63FRR30 pKa = 11.84KK31 pKa = 10.01NLPPGCSQKK40 pKa = 10.96GNSPAQCPNGLTLQWNDD57 pKa = 2.91VLVFDD62 pKa = 6.1KK63 pKa = 10.74IAEE66 pKa = 4.27VNVTGNVWLSNAQVFLGANTPLTINATGSISSDD99 pKa = 2.41STMQMFGDD107 pKa = 4.6LNAVGQISLGSDD119 pKa = 3.02NQVQGNLNSQNQITLNNNTDD139 pKa = 3.33VTGSLVAPQVSLVSSNTITGPISATQLTLGFNSQVTGNLNVTNNLSVGSSSSITGSINAGNLQSNSPVSLNGPIVVDD216 pKa = 3.54NEE218 pKa = 3.93FNLASGSTVNGSVSANRR235 pKa = 11.84VTLAPSNSTLTGDD248 pKa = 3.28VTANEE253 pKa = 3.97NVIIGSGNTIDD264 pKa = 4.24GNVAAGFIRR273 pKa = 11.84LEE275 pKa = 4.25ASNATITGNAIATGNIVIDD294 pKa = 3.41WAGQIGGDD302 pKa = 3.34ATAVNITNNAGNTDD316 pKa = 3.91SVGGTAFCDD325 pKa = 3.7TSDD328 pKa = 3.44GAQPLSCQTGSGGGGATQCDD348 pKa = 4.14ALNDD352 pKa = 3.87LVDD355 pKa = 3.61YY356 pKa = 10.85GVIGDD361 pKa = 4.09SGFTYY366 pKa = 10.81GNNSEE371 pKa = 4.29INGTEE376 pKa = 3.44IDD378 pKa = 5.1DD379 pKa = 4.58PNNTNGNTPTPTGVVEE395 pKa = 4.11EE396 pKa = 5.0AVVQFPPLTPNVFPSFNGGNSFTNPANLGPGTYY429 pKa = 10.38NEE431 pKa = 4.84INLSGNNASASLTGGGTYY449 pKa = 10.3QIDD452 pKa = 4.19SINFGPQSQTLTLGPGDD469 pKa = 3.61YY470 pKa = 10.26FVRR473 pKa = 11.84TISLNNDD480 pKa = 2.95STIAIAPAGPVRR492 pKa = 11.84IFIEE496 pKa = 4.51DD497 pKa = 4.07SITGGNNLFFNPTGAVQNLMINLYY521 pKa = 10.36DD522 pKa = 3.82GASFEE527 pKa = 4.46IGNYY531 pKa = 8.27NQGSTTYY538 pKa = 9.24TFNGVLYY545 pKa = 10.64APYY548 pKa = 9.43TSNDD552 pKa = 2.55ITFGNNSNLQGAVLSAGTVDD572 pKa = 3.09IGNNTSINFTDD583 pKa = 3.78AVQDD587 pKa = 3.66AVRR590 pKa = 11.84DD591 pKa = 3.78AFGCDD596 pKa = 3.1TEE598 pKa = 4.56VTDD601 pKa = 3.15IHH603 pKa = 7.0HH604 pKa = 6.46YY605 pKa = 10.21RR606 pKa = 11.84IQHH609 pKa = 5.81PLSVVSCVAAPVQVVACLDD628 pKa = 3.47ASCASRR634 pKa = 11.84YY635 pKa = 8.26TDD637 pKa = 3.15ATEE640 pKa = 3.99VALSSSATGSTFANGGVLGFTGGQGTLGLRR670 pKa = 11.84YY671 pKa = 9.72VDD673 pKa = 4.48GGTTTVSLSNGAPAAQNPTEE693 pKa = 4.9CYY695 pKa = 10.35DD696 pKa = 3.65SAGTATTNCEE706 pKa = 3.66IEE708 pKa = 4.2FRR710 pKa = 11.84TAGLIFTANDD720 pKa = 3.76GLSAIPPSFAGTDD733 pKa = 3.8FDD735 pKa = 4.01VLARR739 pKa = 11.84AVEE742 pKa = 4.57TNTQTGACEE751 pKa = 3.77ARR753 pKa = 11.84LTGAQVLEE761 pKa = 4.54LATSCRR767 pKa = 11.84NPLTCQAGQNFIANGANVPLNDD789 pKa = 3.8AGATLSYY796 pKa = 11.31ANVPVTFDD804 pKa = 3.58ANGNAPLVANYY815 pKa = 9.68TDD817 pKa = 3.55VGALRR822 pKa = 11.84LHH824 pKa = 6.09GRR826 pKa = 11.84VNLSEE831 pKa = 4.68APNADD836 pKa = 3.35EE837 pKa = 6.05SVIDD841 pKa = 4.75DD842 pKa = 4.09PAITLTGTSVNDD854 pKa = 3.73FVVKK858 pKa = 9.9PHH860 pKa = 5.69TLRR863 pKa = 11.84LFAVDD868 pKa = 3.78NANQAVSATTSGGSAFATAGDD889 pKa = 4.08NFSLIITAEE898 pKa = 4.04NANGDD903 pKa = 3.48ATPNFGRR910 pKa = 11.84EE911 pKa = 3.82TPAAEE916 pKa = 4.06AQASYY921 pKa = 11.11VQTLYY926 pKa = 10.49PSTATAPASDD936 pKa = 3.88FVGTQGWQLDD946 pKa = 3.94TNRR949 pKa = 11.84AGSLRR954 pKa = 11.84TDD956 pKa = 3.23SARR959 pKa = 11.84WLDD962 pKa = 3.4VGTIEE967 pKa = 5.65LAPGLNGNSYY977 pKa = 10.81LGAGDD982 pKa = 3.63VAAKK986 pKa = 10.36QNATVGRR993 pKa = 11.84FAPFRR998 pKa = 11.84FALNTSAVANSCVAGDD1014 pKa = 3.95FTYY1017 pKa = 10.2MSEE1020 pKa = 4.04PALTVSYY1027 pKa = 10.42EE1028 pKa = 4.03LYY1030 pKa = 10.94AVDD1033 pKa = 3.38SDD1035 pKa = 4.58GNITEE1040 pKa = 5.03NYY1042 pKa = 9.57DD1043 pKa = 3.17SSGYY1047 pKa = 10.59SGTAEE1052 pKa = 3.94VGFAAANLAPADD1064 pKa = 3.76TATDD1068 pKa = 3.74RR1069 pKa = 11.84FADD1072 pKa = 3.84RR1073 pKa = 11.84LLNLAAQQAWTAGVMTFNAANAGFNRR1099 pKa = 11.84HH1100 pKa = 6.21PDD1102 pKa = 3.48GNIDD1106 pKa = 3.81GPFASLQLGLRR1117 pKa = 11.84VLSEE1121 pKa = 3.4IDD1123 pKa = 3.24NRR1125 pKa = 11.84NFAGSDD1131 pKa = 3.23LTLAAVAGAAAPLNGTAQLRR1151 pKa = 11.84YY1152 pKa = 9.96GRR1154 pKa = 11.84LVLEE1158 pKa = 4.12NTYY1161 pKa = 10.78GPEE1164 pKa = 3.99NEE1166 pKa = 4.67DD1167 pKa = 3.74LSVPLYY1173 pKa = 10.99AEE1175 pKa = 4.18YY1176 pKa = 10.6FDD1178 pKa = 3.91GSRR1181 pKa = 11.84FVTHH1185 pKa = 7.11TDD1187 pKa = 3.6DD1188 pKa = 3.83SCSSALVAGLNVIADD1203 pKa = 4.36PNNLTPVAAGSDD1215 pKa = 3.62STLTDD1220 pKa = 3.17GALPFDD1226 pKa = 4.15TLRR1229 pKa = 11.84WLAPGTGNTGEE1240 pKa = 4.72FIYY1243 pKa = 10.41EE1244 pKa = 3.69YY1245 pKa = 10.49DD1246 pKa = 3.41APAWLEE1252 pKa = 3.93FEE1254 pKa = 4.37WRR1256 pKa = 11.84DD1257 pKa = 3.43EE1258 pKa = 4.15AGAMYY1263 pKa = 10.56LDD1265 pKa = 4.02PRR1267 pKa = 11.84AIGGFGQYY1275 pKa = 9.73RR1276 pKa = 11.84GNSRR1280 pKa = 11.84VLFWKK1285 pKa = 10.07EE1286 pKa = 3.54VNN1288 pKa = 3.47
Molecular weight: 132.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094IVG5|A0A094IVG5_9GAMM Uncharacterized protein OS=Pseudidiomarina atlantica OX=1517416 GN=IDAT_01505 PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.05RR3 pKa = 11.84QKK5 pKa = 10.3RR6 pKa = 11.84DD7 pKa = 3.2RR8 pKa = 11.84LEE10 pKa = 3.91RR11 pKa = 11.84AHH13 pKa = 6.72AQGFKK18 pKa = 10.64AGVAGRR24 pKa = 11.84SKK26 pKa = 10.44EE27 pKa = 3.81LCPYY31 pKa = 9.54QNQEE35 pKa = 5.09IRR37 pKa = 11.84SQWLGGWRR45 pKa = 11.84DD46 pKa = 3.4AQEE49 pKa = 3.92ARR51 pKa = 11.84SVGLYY56 pKa = 9.82RR57 pKa = 5.42
MM1 pKa = 7.42KK2 pKa = 10.05RR3 pKa = 11.84QKK5 pKa = 10.3RR6 pKa = 11.84DD7 pKa = 3.2RR8 pKa = 11.84LEE10 pKa = 3.91RR11 pKa = 11.84AHH13 pKa = 6.72AQGFKK18 pKa = 10.64AGVAGRR24 pKa = 11.84SKK26 pKa = 10.44EE27 pKa = 3.81LCPYY31 pKa = 9.54QNQEE35 pKa = 5.09IRR37 pKa = 11.84SQWLGGWRR45 pKa = 11.84DD46 pKa = 3.4AQEE49 pKa = 3.92ARR51 pKa = 11.84SVGLYY56 pKa = 9.82RR57 pKa = 5.42
Molecular weight: 6.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
760291 |
35 |
1613 |
332.0 |
36.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.48 ± 0.059 | 0.867 ± 0.019 |
5.605 ± 0.038 | 6.252 ± 0.047 |
3.935 ± 0.03 | 7.084 ± 0.05 |
2.28 ± 0.026 | 5.432 ± 0.037 |
3.82 ± 0.045 | 10.648 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.023 | 3.455 ± 0.038 |
4.25 ± 0.03 | 5.307 ± 0.056 |
5.641 ± 0.041 | 5.537 ± 0.035 |
5.219 ± 0.032 | 7.34 ± 0.042 |
1.419 ± 0.023 | 2.993 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
