
Sendai virus (strain Hamamatsu) (SeV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Respirovirus; Murine respirovirus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
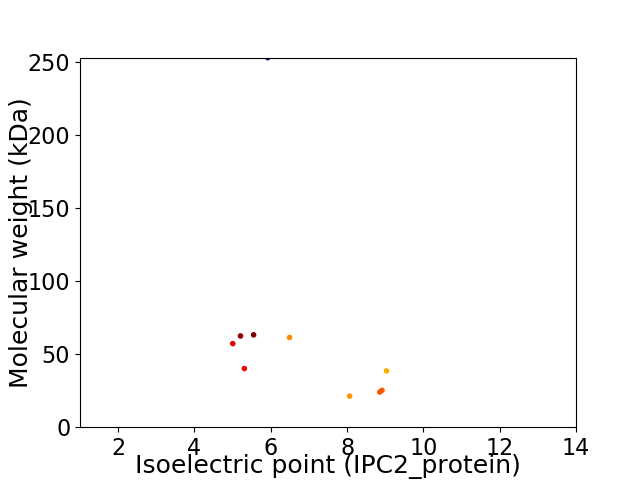
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9DUE0-2|C-2_SENDA Isoform of Q9DUE0 Isoform C of Protein C' OS=Sendai virus (strain Hamamatsu) OX=302271 GN=P/V/C PE=4 SV=3
MM1 pKa = 7.72AGLLSTFDD9 pKa = 3.61TFSSRR14 pKa = 11.84RR15 pKa = 11.84SEE17 pKa = 4.54SINKK21 pKa = 9.79SGGGAVIPGQRR32 pKa = 11.84STVSVFVLGPSVTDD46 pKa = 3.97DD47 pKa = 3.48ADD49 pKa = 3.63KK50 pKa = 11.51LFIATTFLAHH60 pKa = 7.37SLDD63 pKa = 3.34TDD65 pKa = 3.7KK66 pKa = 11.24QHH68 pKa = 5.75SQRR71 pKa = 11.84GGFLVSLLAMAYY83 pKa = 9.77SSPEE87 pKa = 3.87LYY89 pKa = 9.56LTTNGVNADD98 pKa = 3.69VKK100 pKa = 10.02YY101 pKa = 10.81VIYY104 pKa = 10.7NIEE107 pKa = 4.31KK108 pKa = 10.15DD109 pKa = 3.6PKK111 pKa = 8.86RR112 pKa = 11.84TKK114 pKa = 10.08TDD116 pKa = 2.85GFIVKK121 pKa = 8.33TRR123 pKa = 11.84DD124 pKa = 3.0MEE126 pKa = 4.37YY127 pKa = 10.88EE128 pKa = 4.02RR129 pKa = 11.84TTEE132 pKa = 3.92WLFGPMVNKK141 pKa = 10.47SPLFQGQRR149 pKa = 11.84DD150 pKa = 3.65AADD153 pKa = 4.71PDD155 pKa = 4.11TLLQIYY161 pKa = 8.23GYY163 pKa = 7.7PACLGAIIVQVWIVLVKK180 pKa = 10.76AITSSAGLRR189 pKa = 11.84KK190 pKa = 10.19GFFNRR195 pKa = 11.84LEE197 pKa = 4.14AFRR200 pKa = 11.84QDD202 pKa = 3.37GTVKK206 pKa = 10.4GALVFTGEE214 pKa = 4.37TVEE217 pKa = 5.1GIGSVMRR224 pKa = 11.84SQQSLVSLMVEE235 pKa = 4.16TLVTMNTARR244 pKa = 11.84SDD246 pKa = 3.53LTTLEE251 pKa = 4.27KK252 pKa = 10.74NIQIVGNYY260 pKa = 8.94IRR262 pKa = 11.84DD263 pKa = 3.52AGLASFMNTIKK274 pKa = 11.03YY275 pKa = 10.01GVEE278 pKa = 3.61TKK280 pKa = 9.64MAALTLSNLRR290 pKa = 11.84PDD292 pKa = 3.45INKK295 pKa = 9.15LRR297 pKa = 11.84SLIDD301 pKa = 3.3TYY303 pKa = 11.25LSKK306 pKa = 11.09GPRR309 pKa = 11.84APFICILKK317 pKa = 10.47DD318 pKa = 3.39PVHH321 pKa = 7.11GEE323 pKa = 4.29FAPGNYY329 pKa = 7.59PALWSYY335 pKa = 11.87AMGVAVVQNKK345 pKa = 9.23AMQQYY350 pKa = 7.92VTGRR354 pKa = 11.84TYY356 pKa = 11.73LDD358 pKa = 3.07MEE360 pKa = 4.5MFLLGQAVAKK370 pKa = 10.25DD371 pKa = 3.9AEE373 pKa = 4.71SKK375 pKa = 10.5ISSALEE381 pKa = 4.16DD382 pKa = 3.8EE383 pKa = 5.42LGVTDD388 pKa = 3.67TAKK391 pKa = 10.49EE392 pKa = 3.98RR393 pKa = 11.84LRR395 pKa = 11.84HH396 pKa = 5.46HH397 pKa = 7.16LANLSGGDD405 pKa = 3.73GAYY408 pKa = 10.07HH409 pKa = 6.96KK410 pKa = 8.27PTGGGAIEE418 pKa = 4.23VALDD422 pKa = 3.43NADD425 pKa = 3.3IDD427 pKa = 4.61LEE429 pKa = 4.57PEE431 pKa = 3.58AHH433 pKa = 6.61TDD435 pKa = 2.8QDD437 pKa = 3.45ARR439 pKa = 11.84GWGGDD444 pKa = 3.45SGDD447 pKa = 2.92RR448 pKa = 11.84WARR451 pKa = 11.84STSSGHH457 pKa = 7.13FITLHH462 pKa = 4.9GAEE465 pKa = 4.1RR466 pKa = 11.84LEE468 pKa = 4.51EE469 pKa = 4.07EE470 pKa = 4.7TNDD473 pKa = 3.59EE474 pKa = 4.17DD475 pKa = 4.66VSDD478 pKa = 3.67IEE480 pKa = 4.26RR481 pKa = 11.84RR482 pKa = 11.84IARR485 pKa = 11.84RR486 pKa = 11.84LAEE489 pKa = 4.24RR490 pKa = 11.84RR491 pKa = 11.84QEE493 pKa = 3.99DD494 pKa = 3.76ATTHH498 pKa = 5.81EE499 pKa = 4.75DD500 pKa = 3.0EE501 pKa = 4.6GRR503 pKa = 11.84NNGVDD508 pKa = 3.44HH509 pKa = 7.73DD510 pKa = 4.74EE511 pKa = 4.59EE512 pKa = 6.63DD513 pKa = 4.03DD514 pKa = 3.75AAAAAGMGGII524 pKa = 4.58
MM1 pKa = 7.72AGLLSTFDD9 pKa = 3.61TFSSRR14 pKa = 11.84RR15 pKa = 11.84SEE17 pKa = 4.54SINKK21 pKa = 9.79SGGGAVIPGQRR32 pKa = 11.84STVSVFVLGPSVTDD46 pKa = 3.97DD47 pKa = 3.48ADD49 pKa = 3.63KK50 pKa = 11.51LFIATTFLAHH60 pKa = 7.37SLDD63 pKa = 3.34TDD65 pKa = 3.7KK66 pKa = 11.24QHH68 pKa = 5.75SQRR71 pKa = 11.84GGFLVSLLAMAYY83 pKa = 9.77SSPEE87 pKa = 3.87LYY89 pKa = 9.56LTTNGVNADD98 pKa = 3.69VKK100 pKa = 10.02YY101 pKa = 10.81VIYY104 pKa = 10.7NIEE107 pKa = 4.31KK108 pKa = 10.15DD109 pKa = 3.6PKK111 pKa = 8.86RR112 pKa = 11.84TKK114 pKa = 10.08TDD116 pKa = 2.85GFIVKK121 pKa = 8.33TRR123 pKa = 11.84DD124 pKa = 3.0MEE126 pKa = 4.37YY127 pKa = 10.88EE128 pKa = 4.02RR129 pKa = 11.84TTEE132 pKa = 3.92WLFGPMVNKK141 pKa = 10.47SPLFQGQRR149 pKa = 11.84DD150 pKa = 3.65AADD153 pKa = 4.71PDD155 pKa = 4.11TLLQIYY161 pKa = 8.23GYY163 pKa = 7.7PACLGAIIVQVWIVLVKK180 pKa = 10.76AITSSAGLRR189 pKa = 11.84KK190 pKa = 10.19GFFNRR195 pKa = 11.84LEE197 pKa = 4.14AFRR200 pKa = 11.84QDD202 pKa = 3.37GTVKK206 pKa = 10.4GALVFTGEE214 pKa = 4.37TVEE217 pKa = 5.1GIGSVMRR224 pKa = 11.84SQQSLVSLMVEE235 pKa = 4.16TLVTMNTARR244 pKa = 11.84SDD246 pKa = 3.53LTTLEE251 pKa = 4.27KK252 pKa = 10.74NIQIVGNYY260 pKa = 8.94IRR262 pKa = 11.84DD263 pKa = 3.52AGLASFMNTIKK274 pKa = 11.03YY275 pKa = 10.01GVEE278 pKa = 3.61TKK280 pKa = 9.64MAALTLSNLRR290 pKa = 11.84PDD292 pKa = 3.45INKK295 pKa = 9.15LRR297 pKa = 11.84SLIDD301 pKa = 3.3TYY303 pKa = 11.25LSKK306 pKa = 11.09GPRR309 pKa = 11.84APFICILKK317 pKa = 10.47DD318 pKa = 3.39PVHH321 pKa = 7.11GEE323 pKa = 4.29FAPGNYY329 pKa = 7.59PALWSYY335 pKa = 11.87AMGVAVVQNKK345 pKa = 9.23AMQQYY350 pKa = 7.92VTGRR354 pKa = 11.84TYY356 pKa = 11.73LDD358 pKa = 3.07MEE360 pKa = 4.5MFLLGQAVAKK370 pKa = 10.25DD371 pKa = 3.9AEE373 pKa = 4.71SKK375 pKa = 10.5ISSALEE381 pKa = 4.16DD382 pKa = 3.8EE383 pKa = 5.42LGVTDD388 pKa = 3.67TAKK391 pKa = 10.49EE392 pKa = 3.98RR393 pKa = 11.84LRR395 pKa = 11.84HH396 pKa = 5.46HH397 pKa = 7.16LANLSGGDD405 pKa = 3.73GAYY408 pKa = 10.07HH409 pKa = 6.96KK410 pKa = 8.27PTGGGAIEE418 pKa = 4.23VALDD422 pKa = 3.43NADD425 pKa = 3.3IDD427 pKa = 4.61LEE429 pKa = 4.57PEE431 pKa = 3.58AHH433 pKa = 6.61TDD435 pKa = 2.8QDD437 pKa = 3.45ARR439 pKa = 11.84GWGGDD444 pKa = 3.45SGDD447 pKa = 2.92RR448 pKa = 11.84WARR451 pKa = 11.84STSSGHH457 pKa = 7.13FITLHH462 pKa = 4.9GAEE465 pKa = 4.1RR466 pKa = 11.84LEE468 pKa = 4.51EE469 pKa = 4.07EE470 pKa = 4.7TNDD473 pKa = 3.59EE474 pKa = 4.17DD475 pKa = 4.66VSDD478 pKa = 3.67IEE480 pKa = 4.26RR481 pKa = 11.84RR482 pKa = 11.84IARR485 pKa = 11.84RR486 pKa = 11.84LAEE489 pKa = 4.24RR490 pKa = 11.84RR491 pKa = 11.84QEE493 pKa = 3.99DD494 pKa = 3.76ATTHH498 pKa = 5.81EE499 pKa = 4.75DD500 pKa = 3.0EE501 pKa = 4.6GRR503 pKa = 11.84NNGVDD508 pKa = 3.44HH509 pKa = 7.73DD510 pKa = 4.74EE511 pKa = 4.59EE512 pKa = 6.63DD513 pKa = 4.03DD514 pKa = 3.75AAAAAGMGGII524 pKa = 4.58
Molecular weight: 57.2 kDa
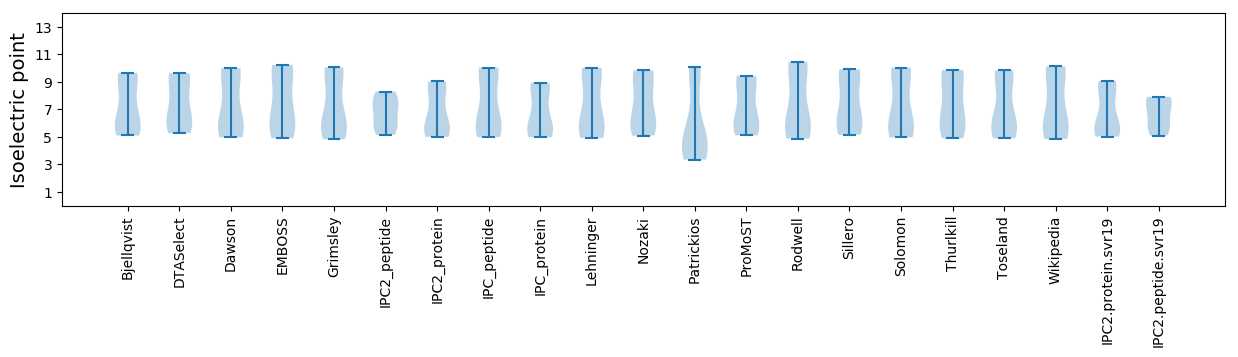
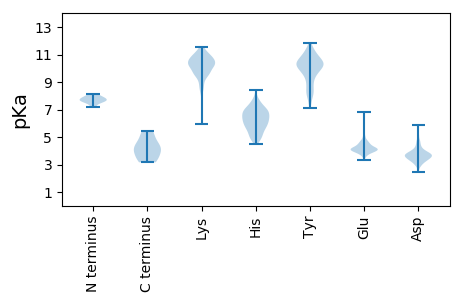
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9DUD8|L_SENDA RNA-directed RNA polymerase L OS=Sendai virus (strain Hamamatsu) OX=302271 GN=L PE=3 SV=1
MM1 pKa = 7.6ADD3 pKa = 3.36IYY5 pKa = 10.85RR6 pKa = 11.84FPKK9 pKa = 10.59FSYY12 pKa = 9.74EE13 pKa = 4.12DD14 pKa = 3.44NGTVEE19 pKa = 4.59PLPLRR24 pKa = 11.84TGPDD28 pKa = 3.01KK29 pKa = 10.99KK30 pKa = 10.56AIPHH34 pKa = 5.1IRR36 pKa = 11.84IVKK39 pKa = 10.01VGDD42 pKa = 3.63PPKK45 pKa = 10.69HH46 pKa = 4.84GVRR49 pKa = 11.84YY50 pKa = 10.03LDD52 pKa = 3.96LLLLGFFEE60 pKa = 4.94TPKK63 pKa = 9.29QTASLGSVSDD73 pKa = 3.85LTEE76 pKa = 3.71HH77 pKa = 6.4TGYY80 pKa = 10.33SICGSGSLPIGVAKK94 pKa = 10.31YY95 pKa = 10.04HH96 pKa = 6.94GSDD99 pKa = 3.34QEE101 pKa = 4.2LLKK104 pKa = 11.05ACTDD108 pKa = 3.21LRR110 pKa = 11.84ITVRR114 pKa = 11.84RR115 pKa = 11.84TVRR118 pKa = 11.84AGEE121 pKa = 4.17MIVYY125 pKa = 8.62MVDD128 pKa = 3.46SIGAPLLPWSGRR140 pKa = 11.84LRR142 pKa = 11.84QGMIFNANKK151 pKa = 9.77VALAPQCLPVDD162 pKa = 3.23KK163 pKa = 10.48DD164 pKa = 2.96IRR166 pKa = 11.84FRR168 pKa = 11.84VVFVNGTSLGAITIAKK184 pKa = 9.25IPKK187 pKa = 8.6TLADD191 pKa = 3.79LALPNSISVNLLVTLKK207 pKa = 10.09TGIPTEE213 pKa = 4.17QKK215 pKa = 10.68GVLPVLDD222 pKa = 4.08DD223 pKa = 3.73QGEE226 pKa = 4.28KK227 pKa = 10.49KK228 pKa = 10.88LNFMVHH234 pKa = 6.52LGLIRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.27VGKK244 pKa = 9.46IYY246 pKa = 10.67SVEE249 pKa = 4.02YY250 pKa = 9.84CKK252 pKa = 11.04SKK254 pKa = 10.18IEE256 pKa = 3.93RR257 pKa = 11.84MRR259 pKa = 11.84LIFSLGLIGGISFHH273 pKa = 5.67VQVTGTLSKK282 pKa = 10.13TFMSQLAWKK291 pKa = 10.01RR292 pKa = 11.84AVCFPLMDD300 pKa = 4.24VNPHH304 pKa = 5.39MNLVIWAASVEE315 pKa = 4.28ITDD318 pKa = 3.38VDD320 pKa = 4.49AVFQPAIPRR329 pKa = 11.84DD330 pKa = 3.38FRR332 pKa = 11.84YY333 pKa = 10.38YY334 pKa = 10.21PNVVAKK340 pKa = 10.68NIGRR344 pKa = 11.84IRR346 pKa = 11.84KK347 pKa = 8.53LL348 pKa = 3.16
MM1 pKa = 7.6ADD3 pKa = 3.36IYY5 pKa = 10.85RR6 pKa = 11.84FPKK9 pKa = 10.59FSYY12 pKa = 9.74EE13 pKa = 4.12DD14 pKa = 3.44NGTVEE19 pKa = 4.59PLPLRR24 pKa = 11.84TGPDD28 pKa = 3.01KK29 pKa = 10.99KK30 pKa = 10.56AIPHH34 pKa = 5.1IRR36 pKa = 11.84IVKK39 pKa = 10.01VGDD42 pKa = 3.63PPKK45 pKa = 10.69HH46 pKa = 4.84GVRR49 pKa = 11.84YY50 pKa = 10.03LDD52 pKa = 3.96LLLLGFFEE60 pKa = 4.94TPKK63 pKa = 9.29QTASLGSVSDD73 pKa = 3.85LTEE76 pKa = 3.71HH77 pKa = 6.4TGYY80 pKa = 10.33SICGSGSLPIGVAKK94 pKa = 10.31YY95 pKa = 10.04HH96 pKa = 6.94GSDD99 pKa = 3.34QEE101 pKa = 4.2LLKK104 pKa = 11.05ACTDD108 pKa = 3.21LRR110 pKa = 11.84ITVRR114 pKa = 11.84RR115 pKa = 11.84TVRR118 pKa = 11.84AGEE121 pKa = 4.17MIVYY125 pKa = 8.62MVDD128 pKa = 3.46SIGAPLLPWSGRR140 pKa = 11.84LRR142 pKa = 11.84QGMIFNANKK151 pKa = 9.77VALAPQCLPVDD162 pKa = 3.23KK163 pKa = 10.48DD164 pKa = 2.96IRR166 pKa = 11.84FRR168 pKa = 11.84VVFVNGTSLGAITIAKK184 pKa = 9.25IPKK187 pKa = 8.6TLADD191 pKa = 3.79LALPNSISVNLLVTLKK207 pKa = 10.09TGIPTEE213 pKa = 4.17QKK215 pKa = 10.68GVLPVLDD222 pKa = 4.08DD223 pKa = 3.73QGEE226 pKa = 4.28KK227 pKa = 10.49KK228 pKa = 10.88LNFMVHH234 pKa = 6.52LGLIRR239 pKa = 11.84RR240 pKa = 11.84KK241 pKa = 9.27VGKK244 pKa = 9.46IYY246 pKa = 10.67SVEE249 pKa = 4.02YY250 pKa = 9.84CKK252 pKa = 11.04SKK254 pKa = 10.18IEE256 pKa = 3.93RR257 pKa = 11.84MRR259 pKa = 11.84LIFSLGLIGGISFHH273 pKa = 5.67VQVTGTLSKK282 pKa = 10.13TFMSQLAWKK291 pKa = 10.01RR292 pKa = 11.84AVCFPLMDD300 pKa = 4.24VNPHH304 pKa = 5.39MNLVIWAASVEE315 pKa = 4.28ITDD318 pKa = 3.38VDD320 pKa = 4.49AVFQPAIPRR329 pKa = 11.84DD330 pKa = 3.38FRR332 pKa = 11.84YY333 pKa = 10.38YY334 pKa = 10.21PNVVAKK340 pKa = 10.68NIGRR344 pKa = 11.84IRR346 pKa = 11.84KK347 pKa = 8.53LL348 pKa = 3.16
Molecular weight: 38.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5777 |
181 |
2228 |
577.7 |
64.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.007 ± 0.58 | 1.489 ± 0.305 |
5.816 ± 0.343 | 6.959 ± 0.717 |
2.51 ± 0.486 | 6.439 ± 0.554 |
1.991 ± 0.089 | 6.612 ± 0.582 |
5.383 ± 0.354 | 9.936 ± 0.889 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.195 | 3.981 ± 0.308 |
4.795 ± 0.532 | 3.566 ± 0.379 |
6.664 ± 0.528 | 8.343 ± 0.51 |
6.699 ± 0.344 | 6.266 ± 0.399 |
1.177 ± 0.196 | 3.185 ± 0.447 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
