
Sanxia tombus-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
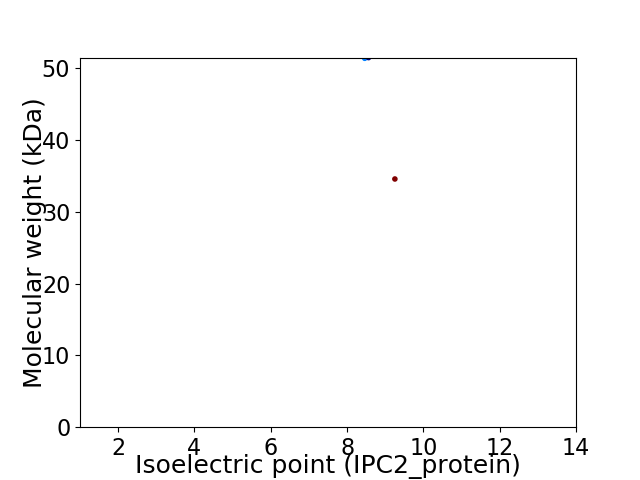
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGI1|A0A1L3KGI1_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 5 OX=1923389 PE=4 SV=1
MM1 pKa = 7.62GSNPIARR8 pKa = 11.84KK9 pKa = 9.41VICHH13 pKa = 5.45QSCVNNEE20 pKa = 3.95IIGLSNRR27 pKa = 11.84HH28 pKa = 5.51IVDD31 pKa = 3.55RR32 pKa = 11.84SYY34 pKa = 11.34IKK36 pKa = 10.26FDD38 pKa = 3.19RR39 pKa = 11.84EE40 pKa = 3.8YY41 pKa = 10.32FLRR44 pKa = 11.84MAKK47 pKa = 8.81ATRR50 pKa = 11.84QEE52 pKa = 4.22LVDD55 pKa = 3.57AAQSEE60 pKa = 4.91KK61 pKa = 10.63VSKK64 pKa = 10.97LNVVYY69 pKa = 9.84GYY71 pKa = 10.86SGGKK75 pKa = 8.48RR76 pKa = 11.84AMYY79 pKa = 10.17LRR81 pKa = 11.84AADD84 pKa = 3.5VPFRR88 pKa = 11.84VAHH91 pKa = 5.56SSITAFVKK99 pKa = 10.33VEE101 pKa = 3.79KK102 pKa = 10.53GYY104 pKa = 11.24EE105 pKa = 3.84EE106 pKa = 6.07DD107 pKa = 3.3IHH109 pKa = 8.06AKK111 pKa = 8.67APRR114 pKa = 11.84MIQYY118 pKa = 9.84RR119 pKa = 11.84SPVYY123 pKa = 10.34NLNLATYY130 pKa = 8.46LKK132 pKa = 9.69PLEE135 pKa = 4.39HH136 pKa = 6.96ALYY139 pKa = 11.04ANLLNEE145 pKa = 4.26NGVPVIAKK153 pKa = 9.09GKK155 pKa = 9.07NPRR158 pKa = 11.84EE159 pKa = 3.86RR160 pKa = 11.84AEE162 pKa = 4.33HH163 pKa = 6.29IKK165 pKa = 10.82ACFDD169 pKa = 3.73DD170 pKa = 4.4KK171 pKa = 11.28PYY173 pKa = 10.96VLAMDD178 pKa = 4.22HH179 pKa = 6.9SKK181 pKa = 10.57FDD183 pKa = 3.21STIRR187 pKa = 11.84VEE189 pKa = 4.23HH190 pKa = 6.38LRR192 pKa = 11.84AEE194 pKa = 4.12HH195 pKa = 6.71KK196 pKa = 10.26IYY198 pKa = 11.14NRR200 pKa = 11.84VFKK203 pKa = 11.04SKK205 pKa = 10.27FLSKK209 pKa = 10.82LLDD212 pKa = 3.64LQIDD216 pKa = 3.56NRR218 pKa = 11.84GYY220 pKa = 8.82TRR222 pKa = 11.84NGLKK226 pKa = 10.3YY227 pKa = 9.27RR228 pKa = 11.84VRR230 pKa = 11.84GTRR233 pKa = 11.84MSGDD237 pKa = 3.5FNTGLGNSVINYY249 pKa = 8.93IVLKK253 pKa = 10.96SFLTMNNIKK262 pKa = 10.28GHH264 pKa = 6.58IYY266 pKa = 10.68LDD268 pKa = 4.16GDD270 pKa = 3.87DD271 pKa = 4.45SLVFIDD277 pKa = 4.71PKK279 pKa = 11.02HH280 pKa = 6.59RR281 pKa = 11.84DD282 pKa = 2.96LDD284 pKa = 4.01FSHH287 pKa = 7.36FEE289 pKa = 3.79KK290 pKa = 10.99CGFEE294 pKa = 4.31TKK296 pKa = 10.14RR297 pKa = 11.84EE298 pKa = 3.98EE299 pKa = 4.36SDD301 pKa = 3.58CLADD305 pKa = 5.03VDD307 pKa = 4.6FCQAKK312 pKa = 10.39YY313 pKa = 9.74MEE315 pKa = 4.75STNLMVRR322 pKa = 11.84NPVRR326 pKa = 11.84AISRR330 pKa = 11.84SLVQVRR336 pKa = 11.84RR337 pKa = 11.84SAFFRR342 pKa = 11.84IRR344 pKa = 11.84AGEE347 pKa = 4.36GIGMTRR353 pKa = 11.84THH355 pKa = 6.93PGVPVLYY362 pKa = 9.62PLYY365 pKa = 10.34KK366 pKa = 10.59QMGVDD371 pKa = 4.0EE372 pKa = 5.13KK373 pKa = 11.2PIFLKK378 pKa = 10.04WYY380 pKa = 8.7PLIDD384 pKa = 3.95DD385 pKa = 4.72RR386 pKa = 11.84EE387 pKa = 4.25VKK389 pKa = 9.18ITDD392 pKa = 3.4AARR395 pKa = 11.84VEE397 pKa = 5.14FWHH400 pKa = 6.59LWGISPDD407 pKa = 3.4EE408 pKa = 3.87QLALEE413 pKa = 4.27EE414 pKa = 4.43SRR416 pKa = 11.84SCGHH420 pKa = 7.02CGTLHH425 pKa = 6.14HH426 pKa = 7.2VEE428 pKa = 4.79FLCPSYY434 pKa = 11.37KK435 pKa = 10.18NAQSVSTTWSGLGG448 pKa = 3.36
MM1 pKa = 7.62GSNPIARR8 pKa = 11.84KK9 pKa = 9.41VICHH13 pKa = 5.45QSCVNNEE20 pKa = 3.95IIGLSNRR27 pKa = 11.84HH28 pKa = 5.51IVDD31 pKa = 3.55RR32 pKa = 11.84SYY34 pKa = 11.34IKK36 pKa = 10.26FDD38 pKa = 3.19RR39 pKa = 11.84EE40 pKa = 3.8YY41 pKa = 10.32FLRR44 pKa = 11.84MAKK47 pKa = 8.81ATRR50 pKa = 11.84QEE52 pKa = 4.22LVDD55 pKa = 3.57AAQSEE60 pKa = 4.91KK61 pKa = 10.63VSKK64 pKa = 10.97LNVVYY69 pKa = 9.84GYY71 pKa = 10.86SGGKK75 pKa = 8.48RR76 pKa = 11.84AMYY79 pKa = 10.17LRR81 pKa = 11.84AADD84 pKa = 3.5VPFRR88 pKa = 11.84VAHH91 pKa = 5.56SSITAFVKK99 pKa = 10.33VEE101 pKa = 3.79KK102 pKa = 10.53GYY104 pKa = 11.24EE105 pKa = 3.84EE106 pKa = 6.07DD107 pKa = 3.3IHH109 pKa = 8.06AKK111 pKa = 8.67APRR114 pKa = 11.84MIQYY118 pKa = 9.84RR119 pKa = 11.84SPVYY123 pKa = 10.34NLNLATYY130 pKa = 8.46LKK132 pKa = 9.69PLEE135 pKa = 4.39HH136 pKa = 6.96ALYY139 pKa = 11.04ANLLNEE145 pKa = 4.26NGVPVIAKK153 pKa = 9.09GKK155 pKa = 9.07NPRR158 pKa = 11.84EE159 pKa = 3.86RR160 pKa = 11.84AEE162 pKa = 4.33HH163 pKa = 6.29IKK165 pKa = 10.82ACFDD169 pKa = 3.73DD170 pKa = 4.4KK171 pKa = 11.28PYY173 pKa = 10.96VLAMDD178 pKa = 4.22HH179 pKa = 6.9SKK181 pKa = 10.57FDD183 pKa = 3.21STIRR187 pKa = 11.84VEE189 pKa = 4.23HH190 pKa = 6.38LRR192 pKa = 11.84AEE194 pKa = 4.12HH195 pKa = 6.71KK196 pKa = 10.26IYY198 pKa = 11.14NRR200 pKa = 11.84VFKK203 pKa = 11.04SKK205 pKa = 10.27FLSKK209 pKa = 10.82LLDD212 pKa = 3.64LQIDD216 pKa = 3.56NRR218 pKa = 11.84GYY220 pKa = 8.82TRR222 pKa = 11.84NGLKK226 pKa = 10.3YY227 pKa = 9.27RR228 pKa = 11.84VRR230 pKa = 11.84GTRR233 pKa = 11.84MSGDD237 pKa = 3.5FNTGLGNSVINYY249 pKa = 8.93IVLKK253 pKa = 10.96SFLTMNNIKK262 pKa = 10.28GHH264 pKa = 6.58IYY266 pKa = 10.68LDD268 pKa = 4.16GDD270 pKa = 3.87DD271 pKa = 4.45SLVFIDD277 pKa = 4.71PKK279 pKa = 11.02HH280 pKa = 6.59RR281 pKa = 11.84DD282 pKa = 2.96LDD284 pKa = 4.01FSHH287 pKa = 7.36FEE289 pKa = 3.79KK290 pKa = 10.99CGFEE294 pKa = 4.31TKK296 pKa = 10.14RR297 pKa = 11.84EE298 pKa = 3.98EE299 pKa = 4.36SDD301 pKa = 3.58CLADD305 pKa = 5.03VDD307 pKa = 4.6FCQAKK312 pKa = 10.39YY313 pKa = 9.74MEE315 pKa = 4.75STNLMVRR322 pKa = 11.84NPVRR326 pKa = 11.84AISRR330 pKa = 11.84SLVQVRR336 pKa = 11.84RR337 pKa = 11.84SAFFRR342 pKa = 11.84IRR344 pKa = 11.84AGEE347 pKa = 4.36GIGMTRR353 pKa = 11.84THH355 pKa = 6.93PGVPVLYY362 pKa = 9.62PLYY365 pKa = 10.34KK366 pKa = 10.59QMGVDD371 pKa = 4.0EE372 pKa = 5.13KK373 pKa = 11.2PIFLKK378 pKa = 10.04WYY380 pKa = 8.7PLIDD384 pKa = 3.95DD385 pKa = 4.72RR386 pKa = 11.84EE387 pKa = 4.25VKK389 pKa = 9.18ITDD392 pKa = 3.4AARR395 pKa = 11.84VEE397 pKa = 5.14FWHH400 pKa = 6.59LWGISPDD407 pKa = 3.4EE408 pKa = 3.87QLALEE413 pKa = 4.27EE414 pKa = 4.43SRR416 pKa = 11.84SCGHH420 pKa = 7.02CGTLHH425 pKa = 6.14HH426 pKa = 7.2VEE428 pKa = 4.79FLCPSYY434 pKa = 11.37KK435 pKa = 10.18NAQSVSTTWSGLGG448 pKa = 3.36
Molecular weight: 51.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI1|A0A1L3KGI1_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 5 OX=1923389 PE=4 SV=1
MM1 pKa = 8.01CIRR4 pKa = 11.84ITMEE8 pKa = 4.61KK9 pKa = 9.99INPTKK14 pKa = 10.18SVRR17 pKa = 11.84RR18 pKa = 11.84AGANVTYY25 pKa = 10.34QRR27 pKa = 11.84NLQRR31 pKa = 11.84LTLSPPAKK39 pKa = 10.22LGVPLGQGPIPFSRR53 pKa = 11.84LGAVLNSGFLPPADD67 pKa = 3.89MLQSVSPKK75 pKa = 9.42RR76 pKa = 11.84DD77 pKa = 3.1KK78 pKa = 11.18PSFKK82 pKa = 10.17KK83 pKa = 9.76DD84 pKa = 3.14TSVQPQATHH93 pKa = 6.44TDD95 pKa = 2.84VTAQKK100 pKa = 10.89KK101 pKa = 5.87MRR103 pKa = 11.84KK104 pKa = 9.04VVITYY109 pKa = 8.93AAKK112 pKa = 9.68PAPPPPAPIVTVPAAKK128 pKa = 10.04PNLATEE134 pKa = 4.13HH135 pKa = 6.28MEE137 pKa = 3.9TRR139 pKa = 11.84LAGHH143 pKa = 6.41RR144 pKa = 11.84QHH146 pKa = 6.31VVKK149 pKa = 10.76GRR151 pKa = 11.84TRR153 pKa = 11.84DD154 pKa = 3.46AYY156 pKa = 11.01CNNIIDD162 pKa = 4.14KK163 pKa = 11.01ADD165 pKa = 3.22IEE167 pKa = 4.5KK168 pKa = 10.35PVLHH172 pKa = 6.32TEE174 pKa = 3.76EE175 pKa = 5.41LIYY178 pKa = 10.42DD179 pKa = 4.17VKK181 pKa = 10.96LLKK184 pKa = 10.35PSFKK188 pKa = 10.62AIVRR192 pKa = 11.84TGVARR197 pKa = 11.84LLRR200 pKa = 11.84AHH202 pKa = 6.13GRR204 pKa = 11.84EE205 pKa = 4.2AIAVEE210 pKa = 4.21AEE212 pKa = 4.3LLGHH216 pKa = 6.72LQLEE220 pKa = 4.43AMFQKK225 pKa = 10.56RR226 pKa = 11.84SAKK229 pKa = 10.35LLLLLKK235 pKa = 9.73TKK237 pKa = 10.57AIQWLKK243 pKa = 10.62NFSKK247 pKa = 10.6IILKK251 pKa = 6.58TTEE254 pKa = 3.3IVALVGRR261 pKa = 11.84TVAAAMCVHH270 pKa = 5.99EE271 pKa = 4.62VEE273 pKa = 4.51EE274 pKa = 4.52EE275 pKa = 3.75ARR277 pKa = 11.84QLLGDD282 pKa = 3.55QAEE285 pKa = 4.33TLNRR289 pKa = 11.84AKK291 pKa = 9.87HH292 pKa = 5.34TDD294 pKa = 3.78FLHH297 pKa = 6.39GKK299 pKa = 8.84CKK301 pKa = 10.5SGLFQSRR308 pKa = 11.84LAGDD312 pKa = 3.85DD313 pKa = 3.34
MM1 pKa = 8.01CIRR4 pKa = 11.84ITMEE8 pKa = 4.61KK9 pKa = 9.99INPTKK14 pKa = 10.18SVRR17 pKa = 11.84RR18 pKa = 11.84AGANVTYY25 pKa = 10.34QRR27 pKa = 11.84NLQRR31 pKa = 11.84LTLSPPAKK39 pKa = 10.22LGVPLGQGPIPFSRR53 pKa = 11.84LGAVLNSGFLPPADD67 pKa = 3.89MLQSVSPKK75 pKa = 9.42RR76 pKa = 11.84DD77 pKa = 3.1KK78 pKa = 11.18PSFKK82 pKa = 10.17KK83 pKa = 9.76DD84 pKa = 3.14TSVQPQATHH93 pKa = 6.44TDD95 pKa = 2.84VTAQKK100 pKa = 10.89KK101 pKa = 5.87MRR103 pKa = 11.84KK104 pKa = 9.04VVITYY109 pKa = 8.93AAKK112 pKa = 9.68PAPPPPAPIVTVPAAKK128 pKa = 10.04PNLATEE134 pKa = 4.13HH135 pKa = 6.28MEE137 pKa = 3.9TRR139 pKa = 11.84LAGHH143 pKa = 6.41RR144 pKa = 11.84QHH146 pKa = 6.31VVKK149 pKa = 10.76GRR151 pKa = 11.84TRR153 pKa = 11.84DD154 pKa = 3.46AYY156 pKa = 11.01CNNIIDD162 pKa = 4.14KK163 pKa = 11.01ADD165 pKa = 3.22IEE167 pKa = 4.5KK168 pKa = 10.35PVLHH172 pKa = 6.32TEE174 pKa = 3.76EE175 pKa = 5.41LIYY178 pKa = 10.42DD179 pKa = 4.17VKK181 pKa = 10.96LLKK184 pKa = 10.35PSFKK188 pKa = 10.62AIVRR192 pKa = 11.84TGVARR197 pKa = 11.84LLRR200 pKa = 11.84AHH202 pKa = 6.13GRR204 pKa = 11.84EE205 pKa = 4.2AIAVEE210 pKa = 4.21AEE212 pKa = 4.3LLGHH216 pKa = 6.72LQLEE220 pKa = 4.43AMFQKK225 pKa = 10.56RR226 pKa = 11.84SAKK229 pKa = 10.35LLLLLKK235 pKa = 9.73TKK237 pKa = 10.57AIQWLKK243 pKa = 10.62NFSKK247 pKa = 10.6IILKK251 pKa = 6.58TTEE254 pKa = 3.3IVALVGRR261 pKa = 11.84TVAAAMCVHH270 pKa = 5.99EE271 pKa = 4.62VEE273 pKa = 4.51EE274 pKa = 4.52EE275 pKa = 3.75ARR277 pKa = 11.84QLLGDD282 pKa = 3.55QAEE285 pKa = 4.33TLNRR289 pKa = 11.84AKK291 pKa = 9.87HH292 pKa = 5.34TDD294 pKa = 3.78FLHH297 pKa = 6.39GKK299 pKa = 8.84CKK301 pKa = 10.5SGLFQSRR308 pKa = 11.84LAGDD312 pKa = 3.85DD313 pKa = 3.34
Molecular weight: 34.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
761 |
313 |
448 |
380.5 |
42.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.279 ± 1.615 | 1.708 ± 0.269 |
4.993 ± 0.725 | 5.519 ± 0.255 |
3.679 ± 0.702 | 5.65 ± 0.337 |
3.548 ± 0.221 | 5.519 ± 0.255 |
7.753 ± 0.746 | 9.593 ± 0.994 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.081 | 4.074 ± 0.749 |
4.993 ± 1.073 | 3.154 ± 0.825 |
7.227 ± 0.324 | 5.782 ± 1.018 |
4.862 ± 1.155 | 7.359 ± 0.007 |
0.657 ± 0.211 | 3.285 ± 1.255 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
