
Halothermothrix orenii (strain H 168 / OCM 544 / DSM 9562)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Halanaerobiales; Halanaerobiaceae; Halothermothrix; Halothermothrix orenii
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
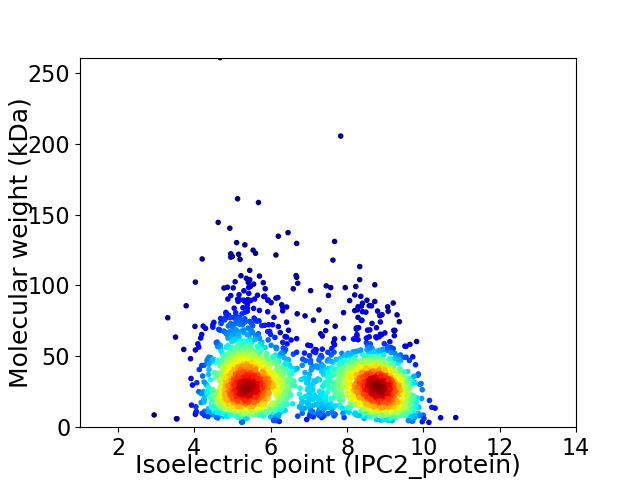
Virtual 2D-PAGE plot for 2324 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8D1W4|B8D1W4_HALOH Radical SAM domain protein OS=Halothermothrix orenii (strain H 168 / OCM 544 / DSM 9562) OX=373903 GN=Hore_04330 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 10.07KK3 pKa = 10.64LSILAILLVGLLVLAGCSKK22 pKa = 10.78DD23 pKa = 4.05ANDD26 pKa = 3.22VNSAVKK32 pKa = 8.83HH33 pKa = 4.98TLTVTVTDD41 pKa = 4.18EE42 pKa = 3.91ATQAAIEE49 pKa = 4.41GATVTVDD56 pKa = 3.42EE57 pKa = 4.86ASKK60 pKa = 7.91TTDD63 pKa = 3.11ANGVAQFEE71 pKa = 4.84LTDD74 pKa = 3.46GTYY77 pKa = 10.44DD78 pKa = 3.09ITVTAEE84 pKa = 4.31GYY86 pKa = 8.48EE87 pKa = 4.32SKK89 pKa = 10.63SGSVTIDD96 pKa = 3.41GKK98 pKa = 10.84DD99 pKa = 3.14STLDD103 pKa = 3.35VTLTAGTGGSTDD115 pKa = 3.69FVLITSNSGEE125 pKa = 4.03EE126 pKa = 3.49TDD128 pKa = 5.63IYY130 pKa = 11.34VDD132 pKa = 3.0WDD134 pKa = 3.49YY135 pKa = 11.84DD136 pKa = 3.87NNIGNIAADD145 pKa = 3.25AWGSGTTITQDD156 pKa = 2.47SSYY159 pKa = 11.36NNTPCWEE166 pKa = 4.16LTTGDD171 pKa = 2.74GWGTVLAFMGDD182 pKa = 3.58IYY184 pKa = 11.44NVDD187 pKa = 3.9QIAEE191 pKa = 4.25FPVDD195 pKa = 3.94LTADD199 pKa = 3.72SVISFSVATTGDD211 pKa = 3.65YY212 pKa = 11.35DD213 pKa = 3.7EE214 pKa = 6.03LRR216 pKa = 11.84VKK218 pKa = 10.63VVGEE222 pKa = 4.04EE223 pKa = 4.06NEE225 pKa = 4.0KK226 pKa = 10.83EE227 pKa = 3.86IAIDD231 pKa = 3.81SFDD234 pKa = 3.62NTSTDD239 pKa = 2.9WQTVQVTTDD248 pKa = 3.16QFSEE252 pKa = 4.47VVPANVTQIAIIAFGGTAGTSKK274 pKa = 10.96VYY276 pKa = 9.4VTDD279 pKa = 3.65YY280 pKa = 9.99TISNAEE286 pKa = 3.9VVIPKK291 pKa = 8.14PQPEE295 pKa = 4.87TYY297 pKa = 9.17TLTVTVTDD305 pKa = 4.41DD306 pKa = 3.49SQVAIEE312 pKa = 4.39GATVTVNGTSKK323 pKa = 7.77TTDD326 pKa = 2.9ASGVATFDD334 pKa = 4.68LPDD337 pKa = 3.17GTYY340 pKa = 9.35TVNVSADD347 pKa = 3.29GYY349 pKa = 10.3ANGSGSVTIDD359 pKa = 3.33GADD362 pKa = 3.26KK363 pKa = 11.15SVDD366 pKa = 3.32VSLTSLEE373 pKa = 4.32VTSTLTVNVKK383 pKa = 10.21NAVSGAVIEE392 pKa = 4.69GAAVTVDD399 pKa = 3.04GTEE402 pKa = 3.64IVTDD406 pKa = 4.11ANGVAQFDD414 pKa = 4.63LVDD417 pKa = 3.72GDD419 pKa = 3.79YY420 pKa = 10.77TINVSANGYY429 pKa = 7.71NTVSQDD435 pKa = 3.24VTIAGADD442 pKa = 3.42KK443 pKa = 10.58TVDD446 pKa = 2.93ISLTTDD452 pKa = 2.96APSLFNSSFTVFNEE466 pKa = 3.9APITALQASYY476 pKa = 10.56EE477 pKa = 4.17VSTTEE482 pKa = 3.46ITEE485 pKa = 4.26GDD487 pKa = 3.55SSIKK491 pKa = 9.0VTYY494 pKa = 10.2PGDD497 pKa = 3.42NWGGIYY503 pKa = 10.29VEE505 pKa = 4.51ISTPVDD511 pKa = 3.48LSSYY515 pKa = 11.09DD516 pKa = 3.32GGNLVFDD523 pKa = 4.43IKK525 pKa = 11.17LPTTIKK531 pKa = 10.75DD532 pKa = 2.95IGVKK536 pKa = 10.56LEE538 pKa = 4.1GPKK541 pKa = 9.69GTGVQVQLANYY552 pKa = 7.46TGTDD556 pKa = 3.33AGNGWMTYY564 pKa = 8.48TIPLSDD570 pKa = 4.65LGIDD574 pKa = 3.95LTQVAVGFGLWNPTDD589 pKa = 3.81GTALDD594 pKa = 3.59AWAGGDD600 pKa = 3.63VYY602 pKa = 10.89IDD604 pKa = 3.37NVRR607 pKa = 11.84FEE609 pKa = 4.25
MM1 pKa = 7.58KK2 pKa = 10.07KK3 pKa = 10.64LSILAILLVGLLVLAGCSKK22 pKa = 10.78DD23 pKa = 4.05ANDD26 pKa = 3.22VNSAVKK32 pKa = 8.83HH33 pKa = 4.98TLTVTVTDD41 pKa = 4.18EE42 pKa = 3.91ATQAAIEE49 pKa = 4.41GATVTVDD56 pKa = 3.42EE57 pKa = 4.86ASKK60 pKa = 7.91TTDD63 pKa = 3.11ANGVAQFEE71 pKa = 4.84LTDD74 pKa = 3.46GTYY77 pKa = 10.44DD78 pKa = 3.09ITVTAEE84 pKa = 4.31GYY86 pKa = 8.48EE87 pKa = 4.32SKK89 pKa = 10.63SGSVTIDD96 pKa = 3.41GKK98 pKa = 10.84DD99 pKa = 3.14STLDD103 pKa = 3.35VTLTAGTGGSTDD115 pKa = 3.69FVLITSNSGEE125 pKa = 4.03EE126 pKa = 3.49TDD128 pKa = 5.63IYY130 pKa = 11.34VDD132 pKa = 3.0WDD134 pKa = 3.49YY135 pKa = 11.84DD136 pKa = 3.87NNIGNIAADD145 pKa = 3.25AWGSGTTITQDD156 pKa = 2.47SSYY159 pKa = 11.36NNTPCWEE166 pKa = 4.16LTTGDD171 pKa = 2.74GWGTVLAFMGDD182 pKa = 3.58IYY184 pKa = 11.44NVDD187 pKa = 3.9QIAEE191 pKa = 4.25FPVDD195 pKa = 3.94LTADD199 pKa = 3.72SVISFSVATTGDD211 pKa = 3.65YY212 pKa = 11.35DD213 pKa = 3.7EE214 pKa = 6.03LRR216 pKa = 11.84VKK218 pKa = 10.63VVGEE222 pKa = 4.04EE223 pKa = 4.06NEE225 pKa = 4.0KK226 pKa = 10.83EE227 pKa = 3.86IAIDD231 pKa = 3.81SFDD234 pKa = 3.62NTSTDD239 pKa = 2.9WQTVQVTTDD248 pKa = 3.16QFSEE252 pKa = 4.47VVPANVTQIAIIAFGGTAGTSKK274 pKa = 10.96VYY276 pKa = 9.4VTDD279 pKa = 3.65YY280 pKa = 9.99TISNAEE286 pKa = 3.9VVIPKK291 pKa = 8.14PQPEE295 pKa = 4.87TYY297 pKa = 9.17TLTVTVTDD305 pKa = 4.41DD306 pKa = 3.49SQVAIEE312 pKa = 4.39GATVTVNGTSKK323 pKa = 7.77TTDD326 pKa = 2.9ASGVATFDD334 pKa = 4.68LPDD337 pKa = 3.17GTYY340 pKa = 9.35TVNVSADD347 pKa = 3.29GYY349 pKa = 10.3ANGSGSVTIDD359 pKa = 3.33GADD362 pKa = 3.26KK363 pKa = 11.15SVDD366 pKa = 3.32VSLTSLEE373 pKa = 4.32VTSTLTVNVKK383 pKa = 10.21NAVSGAVIEE392 pKa = 4.69GAAVTVDD399 pKa = 3.04GTEE402 pKa = 3.64IVTDD406 pKa = 4.11ANGVAQFDD414 pKa = 4.63LVDD417 pKa = 3.72GDD419 pKa = 3.79YY420 pKa = 10.77TINVSANGYY429 pKa = 7.71NTVSQDD435 pKa = 3.24VTIAGADD442 pKa = 3.42KK443 pKa = 10.58TVDD446 pKa = 2.93ISLTTDD452 pKa = 2.96APSLFNSSFTVFNEE466 pKa = 3.9APITALQASYY476 pKa = 10.56EE477 pKa = 4.17VSTTEE482 pKa = 3.46ITEE485 pKa = 4.26GDD487 pKa = 3.55SSIKK491 pKa = 9.0VTYY494 pKa = 10.2PGDD497 pKa = 3.42NWGGIYY503 pKa = 10.29VEE505 pKa = 4.51ISTPVDD511 pKa = 3.48LSSYY515 pKa = 11.09DD516 pKa = 3.32GGNLVFDD523 pKa = 4.43IKK525 pKa = 11.17LPTTIKK531 pKa = 10.75DD532 pKa = 2.95IGVKK536 pKa = 10.56LEE538 pKa = 4.1GPKK541 pKa = 9.69GTGVQVQLANYY552 pKa = 7.46TGTDD556 pKa = 3.33AGNGWMTYY564 pKa = 8.48TIPLSDD570 pKa = 4.65LGIDD574 pKa = 3.95LTQVAVGFGLWNPTDD589 pKa = 3.81GTALDD594 pKa = 3.59AWAGGDD600 pKa = 3.63VYY602 pKa = 10.89IDD604 pKa = 3.37NVRR607 pKa = 11.84FEE609 pKa = 4.25
Molecular weight: 63.57 kDa
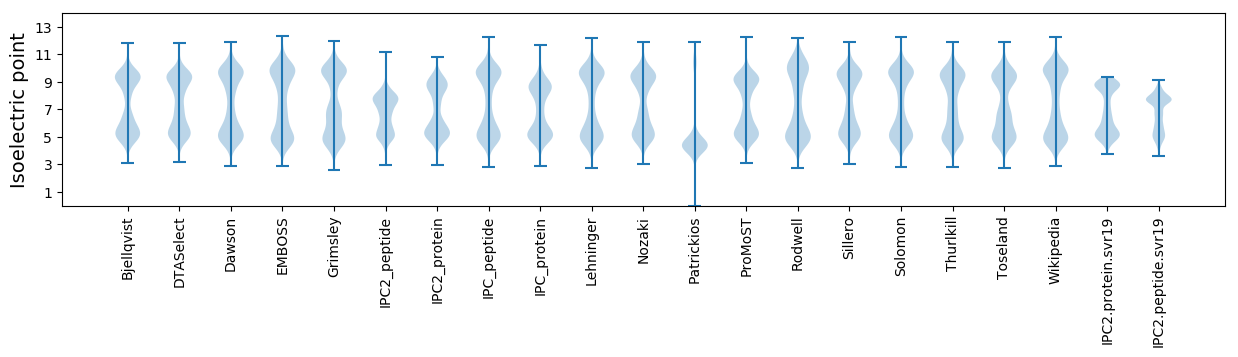
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8CWX6|B8CWX6_HALOH Cytidylate kinase OS=Halothermothrix orenii (strain H 168 / OCM 544 / DSM 9562) OX=373903 GN=cmk PE=3 SV=1
MM1 pKa = 7.62AVPKK5 pKa = 10.44RR6 pKa = 11.84RR7 pKa = 11.84TSKK10 pKa = 9.52ARR12 pKa = 11.84KK13 pKa = 8.45RR14 pKa = 11.84KK15 pKa = 9.53RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 5.94WKK21 pKa = 9.73LKK23 pKa = 10.08SPNLVEE29 pKa = 5.51CPQCHH34 pKa = 5.44EE35 pKa = 4.93LKK37 pKa = 10.51LSHH40 pKa = 6.61RR41 pKa = 11.84VCPSCGYY48 pKa = 8.82YY49 pKa = 10.04KK50 pKa = 10.72GRR52 pKa = 11.84EE53 pKa = 4.22VVSKK57 pKa = 11.13
MM1 pKa = 7.62AVPKK5 pKa = 10.44RR6 pKa = 11.84RR7 pKa = 11.84TSKK10 pKa = 9.52ARR12 pKa = 11.84KK13 pKa = 8.45RR14 pKa = 11.84KK15 pKa = 9.53RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 5.94WKK21 pKa = 9.73LKK23 pKa = 10.08SPNLVEE29 pKa = 5.51CPQCHH34 pKa = 5.44EE35 pKa = 4.93LKK37 pKa = 10.51LSHH40 pKa = 6.61RR41 pKa = 11.84VCPSCGYY48 pKa = 8.82YY49 pKa = 10.04KK50 pKa = 10.72GRR52 pKa = 11.84EE53 pKa = 4.22VVSKK57 pKa = 11.13
Molecular weight: 6.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
736705 |
30 |
2277 |
317.0 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.829 ± 0.052 | 0.732 ± 0.017 |
5.512 ± 0.043 | 7.461 ± 0.053 |
4.144 ± 0.038 | 7.363 ± 0.046 |
1.576 ± 0.018 | 8.794 ± 0.053 |
7.864 ± 0.046 | 9.91 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.343 ± 0.021 | 5.209 ± 0.037 |
3.661 ± 0.025 | 2.661 ± 0.023 |
4.729 ± 0.035 | 5.399 ± 0.035 |
4.836 ± 0.031 | 7.166 ± 0.044 |
0.872 ± 0.022 | 3.94 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
