
marine gamma proteobacterium HTCC2080
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; unclassified Halieaceae
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
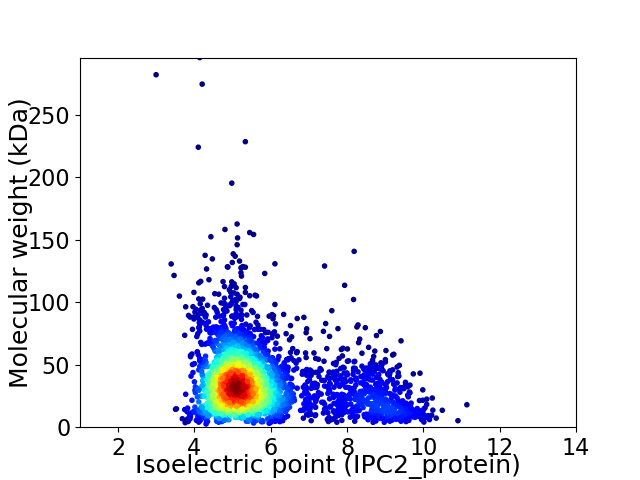
Virtual 2D-PAGE plot for 3184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
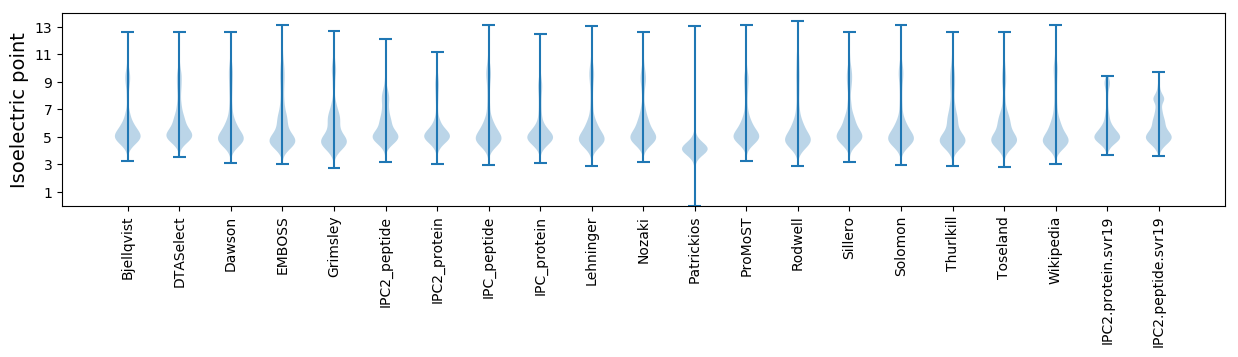
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0Z836|A0Z836_9GAMM HTH hxlR-type domain-containing protein OS=marine gamma proteobacterium HTCC2080 OX=247639 GN=MGP2080_14244 PE=4 SV=1
MM1 pKa = 7.37NNSIIGWGRR10 pKa = 11.84QAARR14 pKa = 11.84VSGFTLTTLSIAISAQASVLEE35 pKa = 4.7EE36 pKa = 3.93IVVTAQKK43 pKa = 10.54RR44 pKa = 11.84EE45 pKa = 3.92QNLQDD50 pKa = 3.29VGVSVTAYY58 pKa = 10.69SGDD61 pKa = 3.35QMKK64 pKa = 10.92ALGVTNTTEE73 pKa = 3.61ITEE76 pKa = 4.29QIAGLQMTSFSPNLVTFNIRR96 pKa = 11.84GVSQNNFTDD105 pKa = 3.59NNEE108 pKa = 3.98APVAVYY114 pKa = 10.34IDD116 pKa = 3.83DD117 pKa = 5.1AYY119 pKa = 9.67VASMNAISGQLFDD132 pKa = 3.81IDD134 pKa = 3.25RR135 pKa = 11.84VEE137 pKa = 4.21VLRR140 pKa = 11.84GPQGTLFGRR149 pKa = 11.84NATGGVIHH157 pKa = 6.45YY158 pKa = 6.91VTKK161 pKa = 10.62GADD164 pKa = 3.12SAEE167 pKa = 3.93FNGYY171 pKa = 9.45VEE173 pKa = 4.66GTYY176 pKa = 10.66SDD178 pKa = 3.76YY179 pKa = 11.47SKK181 pKa = 11.58YY182 pKa = 10.82SIEE185 pKa = 4.19TAFGGSMSANARR197 pKa = 11.84YY198 pKa = 9.73RR199 pKa = 11.84IALRR203 pKa = 11.84KK204 pKa = 9.59EE205 pKa = 3.82EE206 pKa = 4.06SDD208 pKa = 3.99GYY210 pKa = 10.32IEE212 pKa = 4.53SASYY216 pKa = 9.32PQDD219 pKa = 3.34LTVVPVSALPPASGQDD235 pKa = 3.37LGGSNGYY242 pKa = 9.9ALRR245 pKa = 11.84AAFEE249 pKa = 4.4FDD251 pKa = 3.79LSDD254 pKa = 3.68AATLSLSAKK263 pKa = 9.75YY264 pKa = 10.56SEE266 pKa = 5.29DD267 pKa = 3.3SDD269 pKa = 4.49VPTGGYY275 pKa = 10.17SFLPYY280 pKa = 10.21GDD282 pKa = 4.82ASIPPTDD289 pKa = 3.81PADD292 pKa = 3.92PNAYY296 pKa = 9.78VPPEE300 pKa = 3.89FTAFVTDD307 pKa = 4.96IIGAPAGATSAIFFCNDD324 pKa = 3.16QINCFTPVDD333 pKa = 3.67TAGRR337 pKa = 11.84TTFTGDD343 pKa = 2.88SPEE346 pKa = 4.13PFKK349 pKa = 11.32SYY351 pKa = 11.04SDD353 pKa = 3.61YY354 pKa = 11.41AGYY357 pKa = 9.31MDD359 pKa = 6.23RR360 pKa = 11.84DD361 pKa = 4.11TTNLTARR368 pKa = 11.84LDD370 pKa = 3.3WALNDD375 pKa = 3.51NLEE378 pKa = 4.42LVSITNYY385 pKa = 10.35SATDD389 pKa = 3.16KK390 pKa = 10.96FYY392 pKa = 10.86TEE394 pKa = 5.28DD395 pKa = 3.86GDD397 pKa = 5.19GIPAPIIEE405 pKa = 4.73FTTVADD411 pKa = 5.31FSQLSQEE418 pKa = 4.04IRR420 pKa = 11.84LSSEE424 pKa = 3.78GEE426 pKa = 4.13TLRR429 pKa = 11.84WTAGVYY435 pKa = 10.25FLDD438 pKa = 4.1MEE440 pKa = 4.38TDD442 pKa = 3.38ADD444 pKa = 4.28VVTVGAPVGGVAAEE458 pKa = 4.2LGFFDD463 pKa = 5.2GAGNNLAVNARR474 pKa = 11.84VAQDD478 pKa = 3.6YY479 pKa = 11.06LLEE482 pKa = 4.33SQNWSVFGQVEE493 pKa = 4.54LDD495 pKa = 3.27LSDD498 pKa = 4.07RR499 pKa = 11.84LTLIGGYY506 pKa = 9.74RR507 pKa = 11.84YY508 pKa = 10.44SQDD511 pKa = 5.0DD512 pKa = 3.62KK513 pKa = 11.83DD514 pKa = 4.93LDD516 pKa = 3.92FTTTFQADD524 pKa = 3.69GLITDD529 pKa = 4.46VATGATDD536 pKa = 3.91GTAVAVEE543 pKa = 4.05TLNLGNAVAAAGGDD557 pKa = 3.7PQNKK561 pKa = 8.89VDD563 pKa = 3.88YY564 pKa = 10.14SDD566 pKa = 3.55YY567 pKa = 11.12AARR570 pKa = 11.84LQLDD574 pKa = 3.36YY575 pKa = 11.61RR576 pKa = 11.84MNDD579 pKa = 2.84TMLLFASYY587 pKa = 11.14NRR589 pKa = 11.84GIKK592 pKa = 9.98GGNFAPSANVTLEE605 pKa = 4.04QIRR608 pKa = 11.84HH609 pKa = 4.86EE610 pKa = 4.48EE611 pKa = 3.93EE612 pKa = 3.73VLDD615 pKa = 4.17AFEE618 pKa = 4.88VGAKK622 pKa = 9.25TEE624 pKa = 4.13FLDD627 pKa = 4.53GRR629 pKa = 11.84ARR631 pKa = 11.84LNATAFYY638 pKa = 10.52YY639 pKa = 10.35DD640 pKa = 3.62YY641 pKa = 11.53SDD643 pKa = 3.68YY644 pKa = 11.25QAFTFSGGTPSVSNAQAEE662 pKa = 4.41NQGAEE667 pKa = 4.08IEE669 pKa = 4.47LTLLPTEE676 pKa = 4.23NWDD679 pKa = 3.33ILLGVSLQDD688 pKa = 3.36SSVDD692 pKa = 3.51NVEE695 pKa = 4.46TPQSQGTPVGFSVDD709 pKa = 3.26WPVDD713 pKa = 3.62FLNDD717 pKa = 3.36MEE719 pKa = 5.37LPNTPDD725 pKa = 2.73VSFNYY730 pKa = 10.16LFRR733 pKa = 11.84YY734 pKa = 9.6NFDD737 pKa = 3.31VGQGNLAFQFDD748 pKa = 3.8GVYY751 pKa = 10.93YY752 pKa = 10.17GDD754 pKa = 3.84QYY756 pKa = 12.06LEE758 pKa = 4.22VTNGAAAFQKK768 pKa = 10.42SYY770 pKa = 11.09NVSNVSATYY779 pKa = 8.44ATDD782 pKa = 2.95AWSVRR787 pKa = 11.84AWVKK791 pKa = 10.71NVGDD795 pKa = 4.08EE796 pKa = 4.25EE797 pKa = 4.54YY798 pKa = 10.73KK799 pKa = 10.74QYY801 pKa = 11.71ALDD804 pKa = 4.18LGILGGTAVYY814 pKa = 10.11GPPQWWGVTASYY826 pKa = 11.47NFF828 pKa = 3.58
MM1 pKa = 7.37NNSIIGWGRR10 pKa = 11.84QAARR14 pKa = 11.84VSGFTLTTLSIAISAQASVLEE35 pKa = 4.7EE36 pKa = 3.93IVVTAQKK43 pKa = 10.54RR44 pKa = 11.84EE45 pKa = 3.92QNLQDD50 pKa = 3.29VGVSVTAYY58 pKa = 10.69SGDD61 pKa = 3.35QMKK64 pKa = 10.92ALGVTNTTEE73 pKa = 3.61ITEE76 pKa = 4.29QIAGLQMTSFSPNLVTFNIRR96 pKa = 11.84GVSQNNFTDD105 pKa = 3.59NNEE108 pKa = 3.98APVAVYY114 pKa = 10.34IDD116 pKa = 3.83DD117 pKa = 5.1AYY119 pKa = 9.67VASMNAISGQLFDD132 pKa = 3.81IDD134 pKa = 3.25RR135 pKa = 11.84VEE137 pKa = 4.21VLRR140 pKa = 11.84GPQGTLFGRR149 pKa = 11.84NATGGVIHH157 pKa = 6.45YY158 pKa = 6.91VTKK161 pKa = 10.62GADD164 pKa = 3.12SAEE167 pKa = 3.93FNGYY171 pKa = 9.45VEE173 pKa = 4.66GTYY176 pKa = 10.66SDD178 pKa = 3.76YY179 pKa = 11.47SKK181 pKa = 11.58YY182 pKa = 10.82SIEE185 pKa = 4.19TAFGGSMSANARR197 pKa = 11.84YY198 pKa = 9.73RR199 pKa = 11.84IALRR203 pKa = 11.84KK204 pKa = 9.59EE205 pKa = 3.82EE206 pKa = 4.06SDD208 pKa = 3.99GYY210 pKa = 10.32IEE212 pKa = 4.53SASYY216 pKa = 9.32PQDD219 pKa = 3.34LTVVPVSALPPASGQDD235 pKa = 3.37LGGSNGYY242 pKa = 9.9ALRR245 pKa = 11.84AAFEE249 pKa = 4.4FDD251 pKa = 3.79LSDD254 pKa = 3.68AATLSLSAKK263 pKa = 9.75YY264 pKa = 10.56SEE266 pKa = 5.29DD267 pKa = 3.3SDD269 pKa = 4.49VPTGGYY275 pKa = 10.17SFLPYY280 pKa = 10.21GDD282 pKa = 4.82ASIPPTDD289 pKa = 3.81PADD292 pKa = 3.92PNAYY296 pKa = 9.78VPPEE300 pKa = 3.89FTAFVTDD307 pKa = 4.96IIGAPAGATSAIFFCNDD324 pKa = 3.16QINCFTPVDD333 pKa = 3.67TAGRR337 pKa = 11.84TTFTGDD343 pKa = 2.88SPEE346 pKa = 4.13PFKK349 pKa = 11.32SYY351 pKa = 11.04SDD353 pKa = 3.61YY354 pKa = 11.41AGYY357 pKa = 9.31MDD359 pKa = 6.23RR360 pKa = 11.84DD361 pKa = 4.11TTNLTARR368 pKa = 11.84LDD370 pKa = 3.3WALNDD375 pKa = 3.51NLEE378 pKa = 4.42LVSITNYY385 pKa = 10.35SATDD389 pKa = 3.16KK390 pKa = 10.96FYY392 pKa = 10.86TEE394 pKa = 5.28DD395 pKa = 3.86GDD397 pKa = 5.19GIPAPIIEE405 pKa = 4.73FTTVADD411 pKa = 5.31FSQLSQEE418 pKa = 4.04IRR420 pKa = 11.84LSSEE424 pKa = 3.78GEE426 pKa = 4.13TLRR429 pKa = 11.84WTAGVYY435 pKa = 10.25FLDD438 pKa = 4.1MEE440 pKa = 4.38TDD442 pKa = 3.38ADD444 pKa = 4.28VVTVGAPVGGVAAEE458 pKa = 4.2LGFFDD463 pKa = 5.2GAGNNLAVNARR474 pKa = 11.84VAQDD478 pKa = 3.6YY479 pKa = 11.06LLEE482 pKa = 4.33SQNWSVFGQVEE493 pKa = 4.54LDD495 pKa = 3.27LSDD498 pKa = 4.07RR499 pKa = 11.84LTLIGGYY506 pKa = 9.74RR507 pKa = 11.84YY508 pKa = 10.44SQDD511 pKa = 5.0DD512 pKa = 3.62KK513 pKa = 11.83DD514 pKa = 4.93LDD516 pKa = 3.92FTTTFQADD524 pKa = 3.69GLITDD529 pKa = 4.46VATGATDD536 pKa = 3.91GTAVAVEE543 pKa = 4.05TLNLGNAVAAAGGDD557 pKa = 3.7PQNKK561 pKa = 8.89VDD563 pKa = 3.88YY564 pKa = 10.14SDD566 pKa = 3.55YY567 pKa = 11.12AARR570 pKa = 11.84LQLDD574 pKa = 3.36YY575 pKa = 11.61RR576 pKa = 11.84MNDD579 pKa = 2.84TMLLFASYY587 pKa = 11.14NRR589 pKa = 11.84GIKK592 pKa = 9.98GGNFAPSANVTLEE605 pKa = 4.04QIRR608 pKa = 11.84HH609 pKa = 4.86EE610 pKa = 4.48EE611 pKa = 3.93EE612 pKa = 3.73VLDD615 pKa = 4.17AFEE618 pKa = 4.88VGAKK622 pKa = 9.25TEE624 pKa = 4.13FLDD627 pKa = 4.53GRR629 pKa = 11.84ARR631 pKa = 11.84LNATAFYY638 pKa = 10.52YY639 pKa = 10.35DD640 pKa = 3.62YY641 pKa = 11.53SDD643 pKa = 3.68YY644 pKa = 11.25QAFTFSGGTPSVSNAQAEE662 pKa = 4.41NQGAEE667 pKa = 4.08IEE669 pKa = 4.47LTLLPTEE676 pKa = 4.23NWDD679 pKa = 3.33ILLGVSLQDD688 pKa = 3.36SSVDD692 pKa = 3.51NVEE695 pKa = 4.46TPQSQGTPVGFSVDD709 pKa = 3.26WPVDD713 pKa = 3.62FLNDD717 pKa = 3.36MEE719 pKa = 5.37LPNTPDD725 pKa = 2.73VSFNYY730 pKa = 10.16LFRR733 pKa = 11.84YY734 pKa = 9.6NFDD737 pKa = 3.31VGQGNLAFQFDD748 pKa = 3.8GVYY751 pKa = 10.93YY752 pKa = 10.17GDD754 pKa = 3.84QYY756 pKa = 12.06LEE758 pKa = 4.22VTNGAAAFQKK768 pKa = 10.42SYY770 pKa = 11.09NVSNVSATYY779 pKa = 8.44ATDD782 pKa = 2.95AWSVRR787 pKa = 11.84AWVKK791 pKa = 10.71NVGDD795 pKa = 4.08EE796 pKa = 4.25EE797 pKa = 4.54YY798 pKa = 10.73KK799 pKa = 10.74QYY801 pKa = 11.71ALDD804 pKa = 4.18LGILGGTAVYY814 pKa = 10.11GPPQWWGVTASYY826 pKa = 11.47NFF828 pKa = 3.58
Molecular weight: 89.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0Z312|A0Z312_9GAMM Probable transmembrane protein OS=marine gamma proteobacterium HTCC2080 OX=247639 GN=MGP2080_03635 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37SGRR28 pKa = 11.84QVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.98KK41 pKa = 10.26LACC44 pKa = 4.44
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.87RR14 pKa = 11.84THH16 pKa = 5.99GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37SGRR28 pKa = 11.84QVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.98KK41 pKa = 10.26LACC44 pKa = 4.44
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074267 |
24 |
2825 |
337.4 |
36.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.473 ± 0.052 | 1.032 ± 0.014 |
5.857 ± 0.038 | 6.08 ± 0.035 |
3.805 ± 0.027 | 8.078 ± 0.041 |
2.108 ± 0.023 | 5.31 ± 0.026 |
3.311 ± 0.03 | 10.4 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.023 | 3.368 ± 0.023 |
4.67 ± 0.029 | 3.981 ± 0.025 |
5.74 ± 0.037 | 6.424 ± 0.037 |
5.59 ± 0.038 | 7.244 ± 0.036 |
1.467 ± 0.018 | 2.627 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
