
Felis domesticus papillomavirus type 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Lambdapapillomavirus; Lambdapapillomavirus 1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
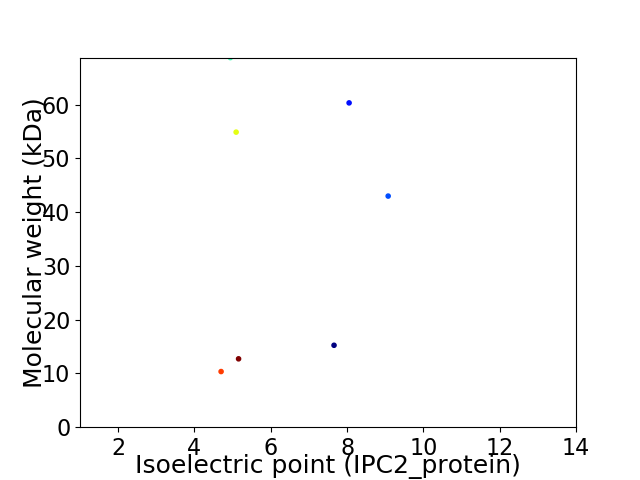
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
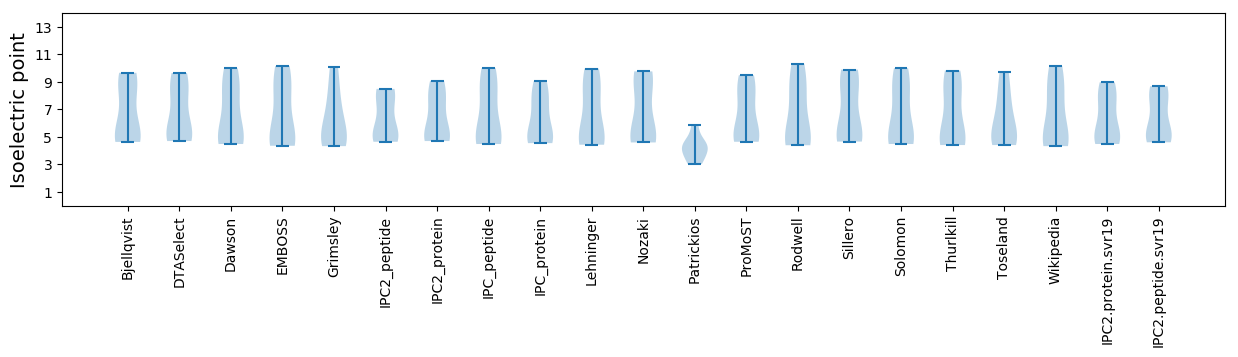
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q77DW1|Q77DW1_9PAPI Protein E6 OS=Felis domesticus papillomavirus type 1 OX=188524 GN=E6 PE=3 SV=1
MM1 pKa = 7.65IGQAPTISDD10 pKa = 3.15IVLTEE15 pKa = 4.0QPPEE19 pKa = 4.2TVDD22 pKa = 3.44LRR24 pKa = 11.84CYY26 pKa = 9.66EE27 pKa = 3.96QMPGEE32 pKa = 4.14EE33 pKa = 4.63EE34 pKa = 4.12EE35 pKa = 4.73VEE37 pKa = 4.26SQARR41 pKa = 11.84DD42 pKa = 3.59LYY44 pKa = 10.79RR45 pKa = 11.84VSSDD49 pKa = 3.82CGLCGSGVRR58 pKa = 11.84FACLAGGEE66 pKa = 4.74DD67 pKa = 3.66ISHH70 pKa = 6.64LRR72 pKa = 11.84SLLTRR77 pKa = 11.84VQVVCVTCVKK87 pKa = 9.67EE88 pKa = 3.89QKK90 pKa = 10.18LNHH93 pKa = 6.41GGG95 pKa = 3.35
MM1 pKa = 7.65IGQAPTISDD10 pKa = 3.15IVLTEE15 pKa = 4.0QPPEE19 pKa = 4.2TVDD22 pKa = 3.44LRR24 pKa = 11.84CYY26 pKa = 9.66EE27 pKa = 3.96QMPGEE32 pKa = 4.14EE33 pKa = 4.63EE34 pKa = 4.12EE35 pKa = 4.73VEE37 pKa = 4.26SQARR41 pKa = 11.84DD42 pKa = 3.59LYY44 pKa = 10.79RR45 pKa = 11.84VSSDD49 pKa = 3.82CGLCGSGVRR58 pKa = 11.84FACLAGGEE66 pKa = 4.74DD67 pKa = 3.66ISHH70 pKa = 6.64LRR72 pKa = 11.84SLLTRR77 pKa = 11.84VQVVCVTCVKK87 pKa = 9.67EE88 pKa = 3.89QKK90 pKa = 10.18LNHH93 pKa = 6.41GGG95 pKa = 3.35
Molecular weight: 10.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q77DV9|Q77DV9_9PAPI Replication protein E1 OS=Felis domesticus papillomavirus type 1 OX=188524 GN=E1 PE=3 SV=1
MM1 pKa = 7.47EE2 pKa = 5.92NISKK6 pKa = 10.56ALDD9 pKa = 4.03SIQEE13 pKa = 4.04QLLTLYY19 pKa = 10.31EE20 pKa = 4.58KK21 pKa = 10.85DD22 pKa = 3.71SADD25 pKa = 3.31LTDD28 pKa = 5.58QIEE31 pKa = 4.24HH32 pKa = 4.94WHH34 pKa = 5.86LNRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.89QTLYY43 pKa = 10.62HH44 pKa = 5.48YY45 pKa = 10.8ARR47 pKa = 11.84RR48 pKa = 11.84QGWMRR53 pKa = 11.84IGMNPVPSLAASQSKK68 pKa = 10.18AKK70 pKa = 10.43SAIEE74 pKa = 4.03QEE76 pKa = 4.5LLLQSMQKK84 pKa = 10.12SAFARR89 pKa = 11.84EE90 pKa = 4.09PWTLSDD96 pKa = 3.42TSRR99 pKa = 11.84EE100 pKa = 3.95RR101 pKa = 11.84LLTEE105 pKa = 3.87PAYY108 pKa = 10.51CFKK111 pKa = 10.95KK112 pKa = 10.34GGRR115 pKa = 11.84QVEE118 pKa = 4.0VRR120 pKa = 11.84YY121 pKa = 10.95DD122 pKa = 3.45NDD124 pKa = 3.49RR125 pKa = 11.84DD126 pKa = 3.63NTSRR130 pKa = 11.84HH131 pKa = 4.93VLWDD135 pKa = 4.46FIYY138 pKa = 10.1FQGDD142 pKa = 2.75NDD144 pKa = 3.89EE145 pKa = 4.12WHH147 pKa = 6.28KK148 pKa = 9.96TPGRR152 pKa = 11.84LDD154 pKa = 3.41ARR156 pKa = 11.84GLYY159 pKa = 10.64YY160 pKa = 9.73MDD162 pKa = 4.45GKK164 pKa = 11.14VKK166 pKa = 9.7TYY168 pKa = 10.5YY169 pKa = 10.82VDD171 pKa = 4.57FEE173 pKa = 4.56EE174 pKa = 4.52EE175 pKa = 3.68AKK177 pKa = 10.67KK178 pKa = 10.64YY179 pKa = 10.68GKK181 pKa = 8.66TGTYY185 pKa = 10.01EE186 pKa = 3.97ILNKK190 pKa = 9.37LTTPVPTSTSSPTGPRR206 pKa = 11.84DD207 pKa = 3.7SPGSGSAASTGVTPKK222 pKa = 10.38KK223 pKa = 8.96QTPGKK228 pKa = 10.0RR229 pKa = 11.84RR230 pKa = 11.84GPLRR234 pKa = 11.84FTSPKK239 pKa = 9.69GPRR242 pKa = 11.84PGGFRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84GRR252 pKa = 11.84GQGEE256 pKa = 4.46LPAPAPGTITPPSAEE271 pKa = 4.14EE272 pKa = 3.7VGKK275 pKa = 10.66ASEE278 pKa = 4.24TVPRR282 pKa = 11.84GSTTRR287 pKa = 11.84LGRR290 pKa = 11.84LLLDD294 pKa = 3.93ARR296 pKa = 11.84DD297 pKa = 3.86PPLLVLKK304 pKa = 10.58GDD306 pKa = 4.13PNSLKK311 pKa = 10.38CVRR314 pKa = 11.84YY315 pKa = 9.16RR316 pKa = 11.84LKK318 pKa = 10.9GKK320 pKa = 10.35YY321 pKa = 9.63SSLFCWVSTTWSWTASTGTARR342 pKa = 11.84WGGARR347 pKa = 11.84MILTFKK353 pKa = 10.65DD354 pKa = 3.39LEE356 pKa = 4.07QRR358 pKa = 11.84EE359 pKa = 4.54VFVKK363 pKa = 9.37TVKK366 pKa = 10.34LPKK369 pKa = 9.82SVQFFRR375 pKa = 11.84GSFDD379 pKa = 4.19DD380 pKa = 3.99YY381 pKa = 12.03
MM1 pKa = 7.47EE2 pKa = 5.92NISKK6 pKa = 10.56ALDD9 pKa = 4.03SIQEE13 pKa = 4.04QLLTLYY19 pKa = 10.31EE20 pKa = 4.58KK21 pKa = 10.85DD22 pKa = 3.71SADD25 pKa = 3.31LTDD28 pKa = 5.58QIEE31 pKa = 4.24HH32 pKa = 4.94WHH34 pKa = 5.86LNRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 3.89QTLYY43 pKa = 10.62HH44 pKa = 5.48YY45 pKa = 10.8ARR47 pKa = 11.84RR48 pKa = 11.84QGWMRR53 pKa = 11.84IGMNPVPSLAASQSKK68 pKa = 10.18AKK70 pKa = 10.43SAIEE74 pKa = 4.03QEE76 pKa = 4.5LLLQSMQKK84 pKa = 10.12SAFARR89 pKa = 11.84EE90 pKa = 4.09PWTLSDD96 pKa = 3.42TSRR99 pKa = 11.84EE100 pKa = 3.95RR101 pKa = 11.84LLTEE105 pKa = 3.87PAYY108 pKa = 10.51CFKK111 pKa = 10.95KK112 pKa = 10.34GGRR115 pKa = 11.84QVEE118 pKa = 4.0VRR120 pKa = 11.84YY121 pKa = 10.95DD122 pKa = 3.45NDD124 pKa = 3.49RR125 pKa = 11.84DD126 pKa = 3.63NTSRR130 pKa = 11.84HH131 pKa = 4.93VLWDD135 pKa = 4.46FIYY138 pKa = 10.1FQGDD142 pKa = 2.75NDD144 pKa = 3.89EE145 pKa = 4.12WHH147 pKa = 6.28KK148 pKa = 9.96TPGRR152 pKa = 11.84LDD154 pKa = 3.41ARR156 pKa = 11.84GLYY159 pKa = 10.64YY160 pKa = 9.73MDD162 pKa = 4.45GKK164 pKa = 11.14VKK166 pKa = 9.7TYY168 pKa = 10.5YY169 pKa = 10.82VDD171 pKa = 4.57FEE173 pKa = 4.56EE174 pKa = 4.52EE175 pKa = 3.68AKK177 pKa = 10.67KK178 pKa = 10.64YY179 pKa = 10.68GKK181 pKa = 8.66TGTYY185 pKa = 10.01EE186 pKa = 3.97ILNKK190 pKa = 9.37LTTPVPTSTSSPTGPRR206 pKa = 11.84DD207 pKa = 3.7SPGSGSAASTGVTPKK222 pKa = 10.38KK223 pKa = 8.96QTPGKK228 pKa = 10.0RR229 pKa = 11.84RR230 pKa = 11.84GPLRR234 pKa = 11.84FTSPKK239 pKa = 9.69GPRR242 pKa = 11.84PGGFRR247 pKa = 11.84RR248 pKa = 11.84GRR250 pKa = 11.84GRR252 pKa = 11.84GQGEE256 pKa = 4.46LPAPAPGTITPPSAEE271 pKa = 4.14EE272 pKa = 3.7VGKK275 pKa = 10.66ASEE278 pKa = 4.24TVPRR282 pKa = 11.84GSTTRR287 pKa = 11.84LGRR290 pKa = 11.84LLLDD294 pKa = 3.93ARR296 pKa = 11.84DD297 pKa = 3.86PPLLVLKK304 pKa = 10.58GDD306 pKa = 4.13PNSLKK311 pKa = 10.38CVRR314 pKa = 11.84YY315 pKa = 9.16RR316 pKa = 11.84LKK318 pKa = 10.9GKK320 pKa = 10.35YY321 pKa = 9.63SSLFCWVSTTWSWTASTGTARR342 pKa = 11.84WGGARR347 pKa = 11.84MILTFKK353 pKa = 10.65DD354 pKa = 3.39LEE356 pKa = 4.07QRR358 pKa = 11.84EE359 pKa = 4.54VFVKK363 pKa = 9.37TVKK366 pKa = 10.34LPKK369 pKa = 9.82SVQFFRR375 pKa = 11.84GSFDD379 pKa = 4.19DD380 pKa = 3.99YY381 pKa = 12.03
Molecular weight: 43.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2370 |
95 |
607 |
338.6 |
37.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.624 ± 0.775 | 2.321 ± 0.823 |
6.16 ± 0.522 | 6.329 ± 0.407 |
4.43 ± 0.54 | 6.793 ± 0.687 |
1.392 ± 0.27 | 3.713 ± 0.601 |
5.359 ± 0.841 | 9.367 ± 0.64 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.603 ± 0.289 | 2.869 ± 0.801 |
6.92 ± 1.293 | 5.148 ± 0.295 |
6.624 ± 0.833 | 7.215 ± 0.785 |
6.582 ± 0.81 | 6.16 ± 0.808 |
1.35 ± 0.403 | 3.038 ± 0.374 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
