
Marco virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Hapavirus; Marco hapavirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
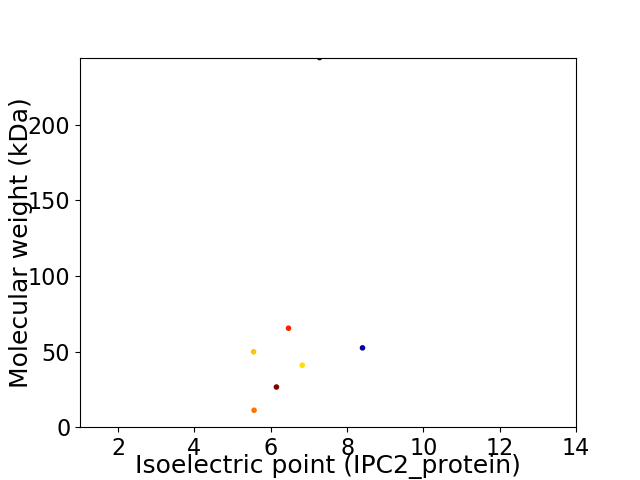
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R1Q9|A0A0D3R1Q9_9RHAB Uncharacterized protein OS=Marco virus OX=1158190 PE=4 SV=1
MM1 pKa = 7.6GSCCGKK7 pKa = 9.63PSRR10 pKa = 11.84RR11 pKa = 11.84SHH13 pKa = 7.31KK14 pKa = 10.27IILPNVLDD22 pKa = 3.5GHH24 pKa = 6.04QFLTDD29 pKa = 3.28EE30 pKa = 4.24SRR32 pKa = 11.84IYY34 pKa = 9.38LTPQDD39 pKa = 4.64PPVSICSVIGHH50 pKa = 7.28DD51 pKa = 3.77ISHH54 pKa = 6.21NRR56 pKa = 11.84NPSVYY61 pKa = 9.56LNHH64 pKa = 6.06YY65 pKa = 9.09NKK67 pKa = 10.77YY68 pKa = 8.95YY69 pKa = 10.84DD70 pKa = 3.5NYY72 pKa = 10.36QNRR75 pKa = 11.84EE76 pKa = 4.06TIRR79 pKa = 11.84YY80 pKa = 9.0IDD82 pKa = 4.74DD83 pKa = 3.93SDD85 pKa = 5.73DD86 pKa = 3.84DD87 pKa = 5.54VYY89 pKa = 11.82DD90 pKa = 3.71QLAATLKK97 pKa = 10.69
MM1 pKa = 7.6GSCCGKK7 pKa = 9.63PSRR10 pKa = 11.84RR11 pKa = 11.84SHH13 pKa = 7.31KK14 pKa = 10.27IILPNVLDD22 pKa = 3.5GHH24 pKa = 6.04QFLTDD29 pKa = 3.28EE30 pKa = 4.24SRR32 pKa = 11.84IYY34 pKa = 9.38LTPQDD39 pKa = 4.64PPVSICSVIGHH50 pKa = 7.28DD51 pKa = 3.77ISHH54 pKa = 6.21NRR56 pKa = 11.84NPSVYY61 pKa = 9.56LNHH64 pKa = 6.06YY65 pKa = 9.09NKK67 pKa = 10.77YY68 pKa = 8.95YY69 pKa = 10.84DD70 pKa = 3.5NYY72 pKa = 10.36QNRR75 pKa = 11.84EE76 pKa = 4.06TIRR79 pKa = 11.84YY80 pKa = 9.0IDD82 pKa = 4.74DD83 pKa = 3.93SDD85 pKa = 5.73DD86 pKa = 3.84DD87 pKa = 5.54VYY89 pKa = 11.82DD90 pKa = 3.71QLAATLKK97 pKa = 10.69
Molecular weight: 11.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R1U6|A0A0D3R1U6_9RHAB GDP polyribonucleotidyltransferase OS=Marco virus OX=1158190 PE=4 SV=1
MM1 pKa = 6.94YY2 pKa = 10.64LKK4 pKa = 9.82TLVLLCGIGVIQSVTQDD21 pKa = 3.35DD22 pKa = 3.98RR23 pKa = 11.84RR24 pKa = 11.84SKK26 pKa = 10.91INLLPFLKK34 pKa = 10.85LEE36 pKa = 4.12WLDD39 pKa = 4.01DD40 pKa = 3.83LRR42 pKa = 11.84KK43 pKa = 9.14PVQDD47 pKa = 3.23VVMDD51 pKa = 3.81SQAVFKK57 pKa = 10.61EE58 pKa = 3.6IFPNILRR65 pKa = 11.84EE66 pKa = 3.96MQAIFQKK73 pKa = 9.99IDD75 pKa = 3.28RR76 pKa = 11.84EE77 pKa = 4.12ISTKK81 pKa = 10.62LFDD84 pKa = 4.32PSRR87 pKa = 11.84EE88 pKa = 4.05AFQKK92 pKa = 10.29MIEE95 pKa = 4.21GTDD98 pKa = 3.71RR99 pKa = 11.84IIKK102 pKa = 8.09QTDD105 pKa = 3.11EE106 pKa = 4.92LFTKK110 pKa = 10.47KK111 pKa = 10.13IPEE114 pKa = 4.42EE115 pKa = 3.87FNKK118 pKa = 9.46MSRR121 pKa = 11.84EE122 pKa = 3.98IARR125 pKa = 11.84STQEE129 pKa = 3.94SINMYY134 pKa = 10.6KK135 pKa = 10.37RR136 pKa = 11.84FEE138 pKa = 4.32KK139 pKa = 10.67DD140 pKa = 3.12FKK142 pKa = 11.47DD143 pKa = 4.01KK144 pKa = 10.63IIEE147 pKa = 4.64PGSHH151 pKa = 7.19AIGSVRR157 pKa = 11.84QEE159 pKa = 3.74ITSSVLMLKK168 pKa = 10.76NNMSTSISRR177 pKa = 11.84MTWNIGNEE185 pKa = 3.95IKK187 pKa = 10.31SLKK190 pKa = 9.32MGLVNEE196 pKa = 5.21KK197 pKa = 9.99ILSLGDD203 pKa = 3.55QINEE207 pKa = 4.03KK208 pKa = 10.33VIKK211 pKa = 9.64KK212 pKa = 8.15TDD214 pKa = 3.32QVLTIFKK221 pKa = 8.77DD222 pKa = 3.4TLNNKK227 pKa = 8.69VSPLFQKK234 pKa = 10.82LSQIDD239 pKa = 3.62FKK241 pKa = 11.71GSVDD245 pKa = 4.15QVNQLIAATKK255 pKa = 9.13GNVVTPVTVLIQSVKK270 pKa = 10.39QLIEE274 pKa = 4.26NYY276 pKa = 10.02SKK278 pKa = 11.1SINNYY283 pKa = 10.03LMPTATKK290 pKa = 10.42ILDD293 pKa = 3.6QVSDD297 pKa = 3.63GTKK300 pKa = 9.53TVFDD304 pKa = 4.5LMSTSSKK311 pKa = 10.2KK312 pKa = 9.67VHH314 pKa = 6.75DD315 pKa = 5.53CILEE319 pKa = 4.07IQASLSTKK327 pKa = 9.68VIPVFEE333 pKa = 4.38GFANYY338 pKa = 7.44VTAISVALKK347 pKa = 10.18PVLEE351 pKa = 4.05NWKK354 pKa = 10.74DD355 pKa = 3.68SIEE358 pKa = 3.84QAFYY362 pKa = 10.6IIGYY366 pKa = 9.34NINQVINQFNIYY378 pKa = 10.31FKK380 pKa = 10.64DD381 pKa = 3.57INWNSWIQNTKK392 pKa = 7.0TWITEE397 pKa = 4.16TYY399 pKa = 10.41LSGKK403 pKa = 9.85AALFDD408 pKa = 4.83FIKK411 pKa = 10.72SASEE415 pKa = 3.88YY416 pKa = 9.91SFKK419 pKa = 10.67AIKK422 pKa = 9.59VIKK425 pKa = 10.02IIAIVLTIITLGIVVIWIINKK446 pKa = 9.07VMIISRR452 pKa = 11.84WCCPNNSS459 pKa = 3.47
MM1 pKa = 6.94YY2 pKa = 10.64LKK4 pKa = 9.82TLVLLCGIGVIQSVTQDD21 pKa = 3.35DD22 pKa = 3.98RR23 pKa = 11.84RR24 pKa = 11.84SKK26 pKa = 10.91INLLPFLKK34 pKa = 10.85LEE36 pKa = 4.12WLDD39 pKa = 4.01DD40 pKa = 3.83LRR42 pKa = 11.84KK43 pKa = 9.14PVQDD47 pKa = 3.23VVMDD51 pKa = 3.81SQAVFKK57 pKa = 10.61EE58 pKa = 3.6IFPNILRR65 pKa = 11.84EE66 pKa = 3.96MQAIFQKK73 pKa = 9.99IDD75 pKa = 3.28RR76 pKa = 11.84EE77 pKa = 4.12ISTKK81 pKa = 10.62LFDD84 pKa = 4.32PSRR87 pKa = 11.84EE88 pKa = 4.05AFQKK92 pKa = 10.29MIEE95 pKa = 4.21GTDD98 pKa = 3.71RR99 pKa = 11.84IIKK102 pKa = 8.09QTDD105 pKa = 3.11EE106 pKa = 4.92LFTKK110 pKa = 10.47KK111 pKa = 10.13IPEE114 pKa = 4.42EE115 pKa = 3.87FNKK118 pKa = 9.46MSRR121 pKa = 11.84EE122 pKa = 3.98IARR125 pKa = 11.84STQEE129 pKa = 3.94SINMYY134 pKa = 10.6KK135 pKa = 10.37RR136 pKa = 11.84FEE138 pKa = 4.32KK139 pKa = 10.67DD140 pKa = 3.12FKK142 pKa = 11.47DD143 pKa = 4.01KK144 pKa = 10.63IIEE147 pKa = 4.64PGSHH151 pKa = 7.19AIGSVRR157 pKa = 11.84QEE159 pKa = 3.74ITSSVLMLKK168 pKa = 10.76NNMSTSISRR177 pKa = 11.84MTWNIGNEE185 pKa = 3.95IKK187 pKa = 10.31SLKK190 pKa = 9.32MGLVNEE196 pKa = 5.21KK197 pKa = 9.99ILSLGDD203 pKa = 3.55QINEE207 pKa = 4.03KK208 pKa = 10.33VIKK211 pKa = 9.64KK212 pKa = 8.15TDD214 pKa = 3.32QVLTIFKK221 pKa = 8.77DD222 pKa = 3.4TLNNKK227 pKa = 8.69VSPLFQKK234 pKa = 10.82LSQIDD239 pKa = 3.62FKK241 pKa = 11.71GSVDD245 pKa = 4.15QVNQLIAATKK255 pKa = 9.13GNVVTPVTVLIQSVKK270 pKa = 10.39QLIEE274 pKa = 4.26NYY276 pKa = 10.02SKK278 pKa = 11.1SINNYY283 pKa = 10.03LMPTATKK290 pKa = 10.42ILDD293 pKa = 3.6QVSDD297 pKa = 3.63GTKK300 pKa = 9.53TVFDD304 pKa = 4.5LMSTSSKK311 pKa = 10.2KK312 pKa = 9.67VHH314 pKa = 6.75DD315 pKa = 5.53CILEE319 pKa = 4.07IQASLSTKK327 pKa = 9.68VIPVFEE333 pKa = 4.38GFANYY338 pKa = 7.44VTAISVALKK347 pKa = 10.18PVLEE351 pKa = 4.05NWKK354 pKa = 10.74DD355 pKa = 3.68SIEE358 pKa = 3.84QAFYY362 pKa = 10.6IIGYY366 pKa = 9.34NINQVINQFNIYY378 pKa = 10.31FKK380 pKa = 10.64DD381 pKa = 3.57INWNSWIQNTKK392 pKa = 7.0TWITEE397 pKa = 4.16TYY399 pKa = 10.41LSGKK403 pKa = 9.85AALFDD408 pKa = 4.83FIKK411 pKa = 10.72SASEE415 pKa = 3.88YY416 pKa = 9.91SFKK419 pKa = 10.67AIKK422 pKa = 9.59VIKK425 pKa = 10.02IIAIVLTIITLGIVVIWIINKK446 pKa = 9.07VMIISRR452 pKa = 11.84WCCPNNSS459 pKa = 3.47
Molecular weight: 52.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4301 |
97 |
2134 |
614.4 |
70.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.022 ± 0.705 | 1.628 ± 0.234 |
5.487 ± 0.446 | 5.929 ± 0.387 |
4.441 ± 0.233 | 5.208 ± 0.667 |
2.232 ± 0.393 | 8.417 ± 0.869 |
7.719 ± 0.828 | 9.579 ± 0.713 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.196 | 5.51 ± 0.409 |
4.743 ± 0.705 | 3.86 ± 0.299 |
3.86 ± 0.38 | 8.603 ± 0.537 |
5.464 ± 0.177 | 5.231 ± 0.521 |
1.883 ± 0.192 | 3.534 ± 0.329 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
