
Apple luteovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
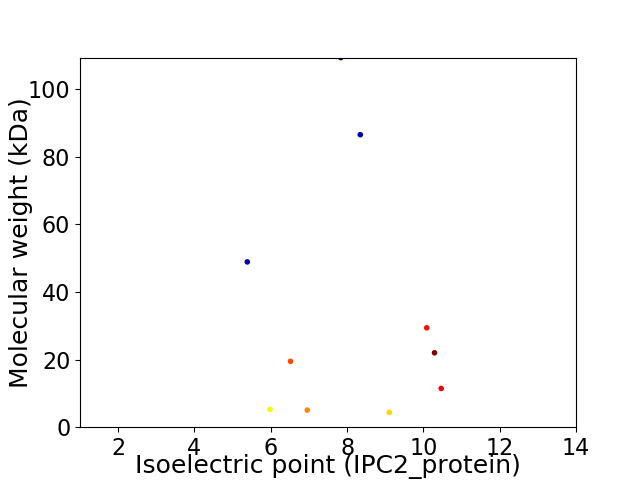
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8N4X5|A0A2U8N4X5_9LUTE RNA-directed RNA polymerase OS=Apple luteovirus 1 OX=2170544 GN=ORF1-ORF2 PE=4 SV=1
MM1 pKa = 7.5LFDD4 pKa = 5.91DD5 pKa = 6.15LICASFKK12 pKa = 10.26VVKK15 pKa = 10.57DD16 pKa = 5.11FISHH20 pKa = 7.01IYY22 pKa = 9.59NNLRR26 pKa = 11.84TVYY29 pKa = 10.48KK30 pKa = 10.4KK31 pKa = 10.57FKK33 pKa = 8.49VWLWEE38 pKa = 3.97LQGKK42 pKa = 9.06FSQHH46 pKa = 6.26DD47 pKa = 3.83AFVDD51 pKa = 3.57ACYY54 pKa = 10.56GYY56 pKa = 9.9MDD58 pKa = 5.35DD59 pKa = 5.06VEE61 pKa = 4.32QFEE64 pKa = 4.58WDD66 pKa = 4.03CYY68 pKa = 10.29SAYY71 pKa = 11.01NDD73 pKa = 4.09ADD75 pKa = 3.71VEE77 pKa = 4.16LALARR82 pKa = 11.84LHH84 pKa = 7.11LDD86 pKa = 3.64TVLKK90 pKa = 10.38APKK93 pKa = 8.19VTGWPVPTRR102 pKa = 11.84PDD104 pKa = 3.43GAPTTTEE111 pKa = 3.91VPKK114 pKa = 11.07YY115 pKa = 7.4PTLHH119 pKa = 6.33EE120 pKa = 4.28LAEE123 pKa = 4.61KK124 pKa = 10.38IRR126 pKa = 11.84TSVRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 3.6RR133 pKa = 11.84VFQAAGEE140 pKa = 4.01ASGDD144 pKa = 3.32KK145 pKa = 10.61DD146 pKa = 3.66AEE148 pKa = 4.42PEE150 pKa = 4.26SEE152 pKa = 4.53VPEE155 pKa = 4.63GWHH158 pKa = 5.06VDD160 pKa = 3.19VWNKK164 pKa = 9.68FQDD167 pKa = 3.51EE168 pKa = 4.24EE169 pKa = 3.96RR170 pKa = 11.84TYY172 pKa = 10.5WEE174 pKa = 4.19NYY176 pKa = 6.99YY177 pKa = 11.23ANPIEE182 pKa = 5.11LAPQVIPVNPGVAEE196 pKa = 4.03PPMPKK201 pKa = 9.55PIYY204 pKa = 7.5TEE206 pKa = 4.12RR207 pKa = 11.84VAFTEE212 pKa = 4.33DD213 pKa = 3.19EE214 pKa = 4.3LFKK217 pKa = 11.42AEE219 pKa = 4.71ARR221 pKa = 11.84LTRR224 pKa = 11.84AKK226 pKa = 10.1RR227 pKa = 11.84SYY229 pKa = 10.98SSTIEE234 pKa = 4.53DD235 pKa = 3.32IKK237 pKa = 11.21DD238 pKa = 3.17QYY240 pKa = 11.33EE241 pKa = 4.06EE242 pKa = 4.38EE243 pKa = 4.19KK244 pKa = 11.24GEE246 pKa = 4.46GYY248 pKa = 10.59FGRR251 pKa = 11.84FFNTFEE257 pKa = 4.01QRR259 pKa = 11.84MHH261 pKa = 5.13YY262 pKa = 9.81VKK264 pKa = 10.57RR265 pKa = 11.84ARR267 pKa = 11.84SRR269 pKa = 11.84RR270 pKa = 11.84AKK272 pKa = 9.74TDD274 pKa = 3.39QLCHH278 pKa = 6.3KK279 pKa = 9.62VQGKK283 pKa = 9.51LSQVAEE289 pKa = 4.16LPDD292 pKa = 4.75FYY294 pKa = 11.11EE295 pKa = 4.27LCTVRR300 pKa = 11.84EE301 pKa = 4.16VEE303 pKa = 3.93TGEE306 pKa = 3.91FHH308 pKa = 6.69TVMDD312 pKa = 4.2EE313 pKa = 4.03GEE315 pKa = 4.28EE316 pKa = 3.87IKK318 pKa = 10.84RR319 pKa = 11.84PIVKK323 pKa = 9.84VSRR326 pKa = 11.84SIKK329 pKa = 9.28PEE331 pKa = 3.73CRR333 pKa = 11.84RR334 pKa = 11.84DD335 pKa = 3.35AQSYY339 pKa = 7.03IRR341 pKa = 11.84KK342 pKa = 9.68YY343 pKa = 10.28IRR345 pKa = 11.84SKK347 pKa = 10.54NNRR350 pKa = 11.84VGADD354 pKa = 3.63EE355 pKa = 5.01IGVATINRR363 pKa = 11.84YY364 pKa = 8.06VAQFADD370 pKa = 4.54DD371 pKa = 4.23MKK373 pKa = 11.55LDD375 pKa = 3.73MASSEE380 pKa = 4.15FLARR384 pKa = 11.84TALTIVPVVTKK395 pKa = 10.69QEE397 pKa = 4.0MVQAMVIHH405 pKa = 6.42SPAARR410 pKa = 11.84KK411 pKa = 9.31ARR413 pKa = 11.84ADD415 pKa = 3.54LAALEE420 pKa = 4.61GQDD423 pKa = 3.58FF424 pKa = 3.83
MM1 pKa = 7.5LFDD4 pKa = 5.91DD5 pKa = 6.15LICASFKK12 pKa = 10.26VVKK15 pKa = 10.57DD16 pKa = 5.11FISHH20 pKa = 7.01IYY22 pKa = 9.59NNLRR26 pKa = 11.84TVYY29 pKa = 10.48KK30 pKa = 10.4KK31 pKa = 10.57FKK33 pKa = 8.49VWLWEE38 pKa = 3.97LQGKK42 pKa = 9.06FSQHH46 pKa = 6.26DD47 pKa = 3.83AFVDD51 pKa = 3.57ACYY54 pKa = 10.56GYY56 pKa = 9.9MDD58 pKa = 5.35DD59 pKa = 5.06VEE61 pKa = 4.32QFEE64 pKa = 4.58WDD66 pKa = 4.03CYY68 pKa = 10.29SAYY71 pKa = 11.01NDD73 pKa = 4.09ADD75 pKa = 3.71VEE77 pKa = 4.16LALARR82 pKa = 11.84LHH84 pKa = 7.11LDD86 pKa = 3.64TVLKK90 pKa = 10.38APKK93 pKa = 8.19VTGWPVPTRR102 pKa = 11.84PDD104 pKa = 3.43GAPTTTEE111 pKa = 3.91VPKK114 pKa = 11.07YY115 pKa = 7.4PTLHH119 pKa = 6.33EE120 pKa = 4.28LAEE123 pKa = 4.61KK124 pKa = 10.38IRR126 pKa = 11.84TSVRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 3.6RR133 pKa = 11.84VFQAAGEE140 pKa = 4.01ASGDD144 pKa = 3.32KK145 pKa = 10.61DD146 pKa = 3.66AEE148 pKa = 4.42PEE150 pKa = 4.26SEE152 pKa = 4.53VPEE155 pKa = 4.63GWHH158 pKa = 5.06VDD160 pKa = 3.19VWNKK164 pKa = 9.68FQDD167 pKa = 3.51EE168 pKa = 4.24EE169 pKa = 3.96RR170 pKa = 11.84TYY172 pKa = 10.5WEE174 pKa = 4.19NYY176 pKa = 6.99YY177 pKa = 11.23ANPIEE182 pKa = 5.11LAPQVIPVNPGVAEE196 pKa = 4.03PPMPKK201 pKa = 9.55PIYY204 pKa = 7.5TEE206 pKa = 4.12RR207 pKa = 11.84VAFTEE212 pKa = 4.33DD213 pKa = 3.19EE214 pKa = 4.3LFKK217 pKa = 11.42AEE219 pKa = 4.71ARR221 pKa = 11.84LTRR224 pKa = 11.84AKK226 pKa = 10.1RR227 pKa = 11.84SYY229 pKa = 10.98SSTIEE234 pKa = 4.53DD235 pKa = 3.32IKK237 pKa = 11.21DD238 pKa = 3.17QYY240 pKa = 11.33EE241 pKa = 4.06EE242 pKa = 4.38EE243 pKa = 4.19KK244 pKa = 11.24GEE246 pKa = 4.46GYY248 pKa = 10.59FGRR251 pKa = 11.84FFNTFEE257 pKa = 4.01QRR259 pKa = 11.84MHH261 pKa = 5.13YY262 pKa = 9.81VKK264 pKa = 10.57RR265 pKa = 11.84ARR267 pKa = 11.84SRR269 pKa = 11.84RR270 pKa = 11.84AKK272 pKa = 9.74TDD274 pKa = 3.39QLCHH278 pKa = 6.3KK279 pKa = 9.62VQGKK283 pKa = 9.51LSQVAEE289 pKa = 4.16LPDD292 pKa = 4.75FYY294 pKa = 11.11EE295 pKa = 4.27LCTVRR300 pKa = 11.84EE301 pKa = 4.16VEE303 pKa = 3.93TGEE306 pKa = 3.91FHH308 pKa = 6.69TVMDD312 pKa = 4.2EE313 pKa = 4.03GEE315 pKa = 4.28EE316 pKa = 3.87IKK318 pKa = 10.84RR319 pKa = 11.84PIVKK323 pKa = 9.84VSRR326 pKa = 11.84SIKK329 pKa = 9.28PEE331 pKa = 3.73CRR333 pKa = 11.84RR334 pKa = 11.84DD335 pKa = 3.35AQSYY339 pKa = 7.03IRR341 pKa = 11.84KK342 pKa = 9.68YY343 pKa = 10.28IRR345 pKa = 11.84SKK347 pKa = 10.54NNRR350 pKa = 11.84VGADD354 pKa = 3.63EE355 pKa = 5.01IGVATINRR363 pKa = 11.84YY364 pKa = 8.06VAQFADD370 pKa = 4.54DD371 pKa = 4.23MKK373 pKa = 11.55LDD375 pKa = 3.73MASSEE380 pKa = 4.15FLARR384 pKa = 11.84TALTIVPVVTKK395 pKa = 10.69QEE397 pKa = 4.0MVQAMVIHH405 pKa = 6.42SPAARR410 pKa = 11.84KK411 pKa = 9.31ARR413 pKa = 11.84ADD415 pKa = 3.54LAALEE420 pKa = 4.61GQDD423 pKa = 3.58FF424 pKa = 3.83
Molecular weight: 48.92 kDa
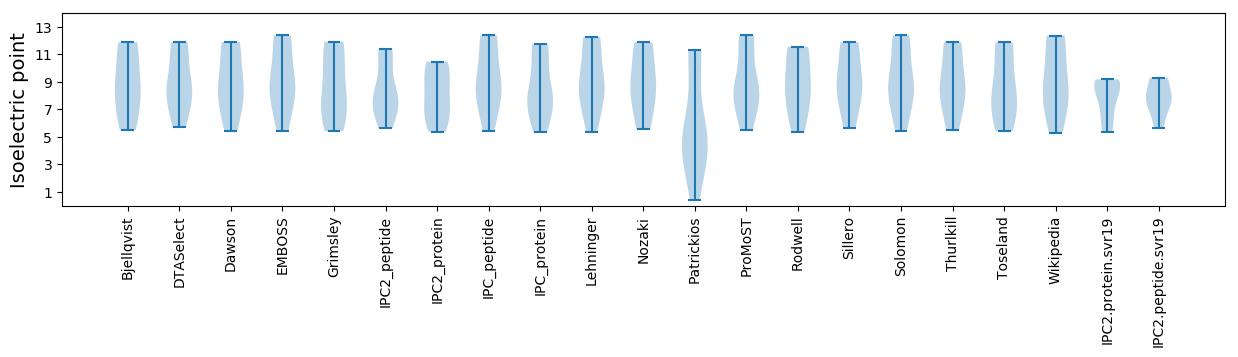
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8N4S3|A0A2U8N4S3_9LUTE Readthrough protein OS=Apple luteovirus 1 OX=2170544 GN=ORF3-ORF5 PE=3 SV=1
MM1 pKa = 7.36TPSSMPVTVTWTTSSSSSGTATQHH25 pKa = 5.61TMMLMLNLPSPDD37 pKa = 3.51STSTPSLRR45 pKa = 11.84LLKK48 pKa = 10.72LPAGPSQQDD57 pKa = 3.14LMVRR61 pKa = 11.84QPRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84SPNTRR71 pKa = 11.84LCTNSQKK78 pKa = 10.59RR79 pKa = 11.84SAQACDD85 pKa = 2.83EE86 pKa = 4.18SGYY89 pKa = 11.07FKK91 pKa = 11.05LQVKK95 pKa = 10.12RR96 pKa = 11.84LVIKK100 pKa = 8.36MQNLSLKK107 pKa = 8.14CQRR110 pKa = 11.84GGTLTSGTSSKK121 pKa = 9.36TRR123 pKa = 11.84SEE125 pKa = 4.14PTGKK129 pKa = 7.33TTTPTPSSWRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84LYY143 pKa = 11.05LSTQGSLNLPCPSRR157 pKa = 11.84YY158 pKa = 7.92TPRR161 pKa = 11.84EE162 pKa = 3.67SPLRR166 pKa = 11.84RR167 pKa = 11.84TSSSKK172 pKa = 10.61LRR174 pKa = 11.84LGSPGQNVHH183 pKa = 5.65TRR185 pKa = 11.84PPLKK189 pKa = 9.94TSRR192 pKa = 11.84TSMKK196 pKa = 9.92KK197 pKa = 8.82RR198 pKa = 11.84RR199 pKa = 11.84EE200 pKa = 4.12RR201 pKa = 11.84ATSAAFLTRR210 pKa = 11.84LSSEE214 pKa = 4.18CIMLRR219 pKa = 11.84EE220 pKa = 4.08RR221 pKa = 11.84GAVEE225 pKa = 3.82PRR227 pKa = 11.84RR228 pKa = 11.84TSCVTRR234 pKa = 11.84FKK236 pKa = 11.11VNSVRR241 pKa = 11.84LLNYY245 pKa = 9.33LISMSCVPSEE255 pKa = 3.44KK256 pKa = 9.56WKK258 pKa = 10.11PVSSTLL264 pKa = 3.99
MM1 pKa = 7.36TPSSMPVTVTWTTSSSSSGTATQHH25 pKa = 5.61TMMLMLNLPSPDD37 pKa = 3.51STSTPSLRR45 pKa = 11.84LLKK48 pKa = 10.72LPAGPSQQDD57 pKa = 3.14LMVRR61 pKa = 11.84QPRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84SPNTRR71 pKa = 11.84LCTNSQKK78 pKa = 10.59RR79 pKa = 11.84SAQACDD85 pKa = 2.83EE86 pKa = 4.18SGYY89 pKa = 11.07FKK91 pKa = 11.05LQVKK95 pKa = 10.12RR96 pKa = 11.84LVIKK100 pKa = 8.36MQNLSLKK107 pKa = 8.14CQRR110 pKa = 11.84GGTLTSGTSSKK121 pKa = 9.36TRR123 pKa = 11.84SEE125 pKa = 4.14PTGKK129 pKa = 7.33TTTPTPSSWRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84LYY143 pKa = 11.05LSTQGSLNLPCPSRR157 pKa = 11.84YY158 pKa = 7.92TPRR161 pKa = 11.84EE162 pKa = 3.67SPLRR166 pKa = 11.84RR167 pKa = 11.84TSSSKK172 pKa = 10.61LRR174 pKa = 11.84LGSPGQNVHH183 pKa = 5.65TRR185 pKa = 11.84PPLKK189 pKa = 9.94TSRR192 pKa = 11.84TSMKK196 pKa = 9.92KK197 pKa = 8.82RR198 pKa = 11.84RR199 pKa = 11.84EE200 pKa = 4.12RR201 pKa = 11.84ATSAAFLTRR210 pKa = 11.84LSSEE214 pKa = 4.18CIMLRR219 pKa = 11.84EE220 pKa = 4.08RR221 pKa = 11.84GAVEE225 pKa = 3.82PRR227 pKa = 11.84RR228 pKa = 11.84TSCVTRR234 pKa = 11.84FKK236 pKa = 11.11VNSVRR241 pKa = 11.84LLNYY245 pKa = 9.33LISMSCVPSEE255 pKa = 3.44KK256 pKa = 9.56WKK258 pKa = 10.11PVSSTLL264 pKa = 3.99
Molecular weight: 29.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3024 |
38 |
953 |
302.4 |
34.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.713 ± 0.694 | 1.72 ± 0.221 |
5.126 ± 0.866 | 6.118 ± 1.057 |
3.935 ± 0.562 | 5.754 ± 0.523 |
1.62 ± 0.247 | 4.497 ± 0.671 |
6.052 ± 0.707 | 7.573 ± 1.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.513 ± 0.484 | 4.134 ± 0.532 |
6.581 ± 0.808 | 3.472 ± 0.231 |
8.366 ± 0.894 | 8.267 ± 1.735 |
5.721 ± 0.893 | 7.044 ± 0.732 |
1.389 ± 0.144 | 3.373 ± 0.455 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
