
Marivirga tractuosa (strain ATCC 23168 / DSM 4126 / NBRC 15989 / NCIMB 1408 / VKM B-1430 / H-43) (Microscilla tractuosa) (Flexibacter tractuosus)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Marivirgaceae; Marivirga; Marivirga tractuosa
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
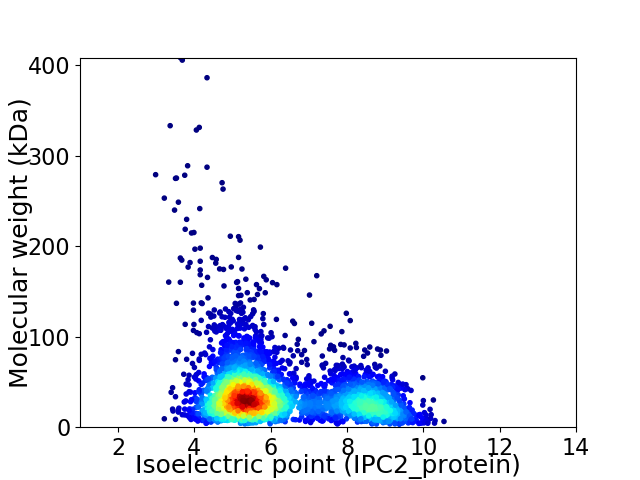
Virtual 2D-PAGE plot for 3748 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
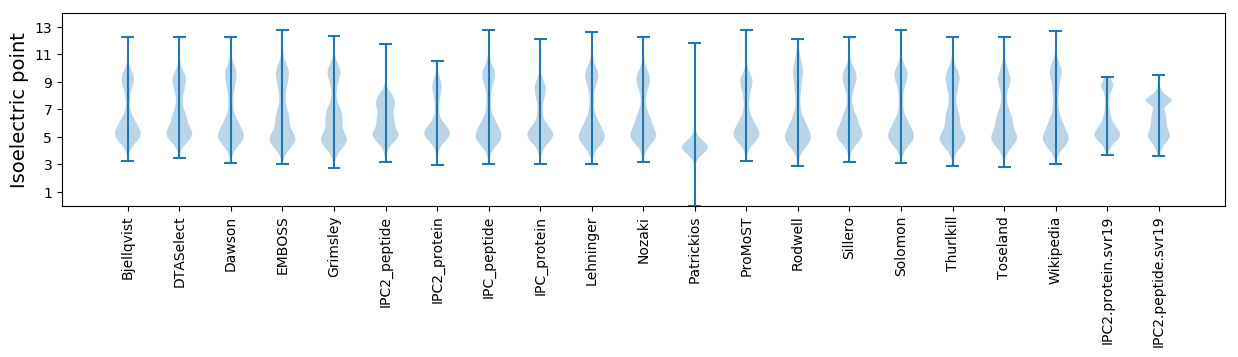
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4TUU0|E4TUU0_MARTH RimK domain protein ATP-grasp OS=Marivirga tractuosa (strain ATCC 23168 / DSM 4126 / NBRC 15989 / NCIMB 1408 / VKM B-1430 / H-43) OX=643867 GN=Ftrac_1048 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 10.38FLPTYY7 pKa = 10.45LFFIGLVFFANQSFGQTSYY26 pKa = 11.07IEE28 pKa = 4.32SVNPKK33 pKa = 9.77NGYY36 pKa = 8.95SGQIINISGVGLAGADD52 pKa = 3.13KK53 pKa = 11.08VFFGSVEE60 pKa = 3.88GKK62 pKa = 10.0IINVEE67 pKa = 3.99DD68 pKa = 3.6QLIEE72 pKa = 4.28AEE74 pKa = 4.58VPAGTTYY81 pKa = 11.68DD82 pKa = 3.77NITVLKK88 pKa = 10.31SSTRR92 pKa = 11.84LSYY95 pKa = 10.85SGEE98 pKa = 4.13HH99 pKa = 6.12FLLSYY104 pKa = 10.67GGEE107 pKa = 4.17QGVSSSDD114 pKa = 3.29FDD116 pKa = 4.11DD117 pKa = 3.97QVDD120 pKa = 3.7VFADD124 pKa = 3.64DD125 pKa = 3.7GLYY128 pKa = 10.86DD129 pKa = 3.68VTLSDD134 pKa = 5.57LDD136 pKa = 4.01GDD138 pKa = 4.25GKK140 pKa = 10.82NDD142 pKa = 3.79IIGANSKK149 pKa = 10.78SNSATILRR157 pKa = 11.84NLSTPSNLSFDD168 pKa = 3.61QRR170 pKa = 11.84NIGIGAPTLNITAGDD185 pKa = 3.97LNGDD189 pKa = 4.23GKK191 pKa = 10.17PDD193 pKa = 3.54VVFSEE198 pKa = 4.79AGDD201 pKa = 3.8GNRR204 pKa = 11.84LIILGNNSSPGSLNFSIQTITLEE227 pKa = 4.3GSSTKK232 pKa = 10.37RR233 pKa = 11.84VVIKK237 pKa = 10.85DD238 pKa = 3.6LDD240 pKa = 4.23LDD242 pKa = 4.09GKK244 pKa = 9.79PDD246 pKa = 3.94LVVSDD251 pKa = 3.65QVNNRR256 pKa = 11.84ILIIKK261 pKa = 7.01NTSSAGTLSFSPEE274 pKa = 3.87IIEE277 pKa = 4.14LTVEE281 pKa = 4.28NAVSTAGLDD290 pKa = 3.76VEE292 pKa = 5.29DD293 pKa = 5.24LNGDD297 pKa = 4.19GKK299 pKa = 10.18PEE301 pKa = 4.73IITHH305 pKa = 5.75QFQTDD310 pKa = 2.85AGGFFIATNQSSPGNFSFTDD330 pKa = 3.69FNKK333 pKa = 10.26YY334 pKa = 7.6STPGTLINLKK344 pKa = 10.6VGDD347 pKa = 4.19INQDD351 pKa = 3.12NKK353 pKa = 10.79PDD355 pKa = 3.4IVATLFLSSSVAVFNNEE372 pKa = 3.7TTDD375 pKa = 3.42TGNVPQFGSAQNLATDD391 pKa = 3.65VRR393 pKa = 11.84PWGLDD398 pKa = 3.33FGDD401 pKa = 4.07MDD403 pKa = 4.88GDD405 pKa = 4.28GIKK408 pKa = 10.45DD409 pKa = 3.22IVVATVGTDD418 pKa = 3.1KK419 pKa = 11.0TINILNNDD427 pKa = 3.19GTGGLNYY434 pKa = 10.06SKK436 pKa = 11.06VSIPVTYY443 pKa = 10.11INRR446 pKa = 11.84NIKK449 pKa = 10.23IGDD452 pKa = 3.35IDD454 pKa = 4.58GDD456 pKa = 4.19SKK458 pKa = 11.36PDD460 pKa = 3.31IVFTSVDD467 pKa = 3.75DD468 pKa = 4.78DD469 pKa = 4.34NNQITASNISILRR482 pKa = 11.84NNQCIIPVITPEE494 pKa = 4.25GPINACEE501 pKa = 4.0GNPVRR506 pKa = 11.84LEE508 pKa = 4.0TQNIEE513 pKa = 3.89GLTFEE518 pKa = 4.78WFQDD522 pKa = 3.41GTSVKK527 pKa = 9.84TGTEE531 pKa = 3.77NFIEE535 pKa = 4.47LNDD538 pKa = 3.57VSATGSYY545 pKa = 8.74TVSIISDD552 pKa = 3.37GGSCQEE558 pKa = 3.35ISEE561 pKa = 4.69AIEE564 pKa = 3.75VNIISEE570 pKa = 4.33GALPSATISSNDD582 pKa = 4.03PICNGGILTLNSSDD596 pKa = 4.89VGATDD601 pKa = 3.64YY602 pKa = 11.03KK603 pKa = 10.17WRR605 pKa = 11.84GPQGYY610 pKa = 6.59TAEE613 pKa = 5.22GITVEE618 pKa = 4.47VDD620 pKa = 3.45DD621 pKa = 5.51FDD623 pKa = 3.89VDD625 pKa = 3.36KK626 pKa = 11.32AGRR629 pKa = 11.84YY630 pKa = 8.86YY631 pKa = 11.22LDD633 pKa = 4.21VYY635 pKa = 11.35SGDD638 pKa = 4.55CIIEE642 pKa = 4.13TTSIVVEE649 pKa = 4.8VIPSPNFSVEE659 pKa = 3.65QSGAGTYY666 pKa = 10.59CEE668 pKa = 4.71GEE670 pKa = 4.36SVTLNVSPSEE680 pKa = 4.06NGFGFQWYY688 pKa = 9.33RR689 pKa = 11.84GTSTISGATSATYY702 pKa = 10.97NPTTDD707 pKa = 2.54GDD709 pKa = 4.64YY710 pKa = 10.97YY711 pKa = 11.65VEE713 pKa = 4.42ITDD716 pKa = 4.58QVNTGCPKK724 pKa = 10.09IYY726 pKa = 10.44SDD728 pKa = 3.67TLQVAFLEE736 pKa = 4.7TPEE739 pKa = 4.31VYY741 pKa = 10.74YY742 pKa = 10.26DD743 pKa = 4.22LPSSACVGIPVSFTNDD759 pKa = 2.5AVVADD764 pKa = 4.67EE765 pKa = 5.0SLAEE769 pKa = 3.94YY770 pKa = 10.2RR771 pKa = 11.84WDD773 pKa = 3.83FGDD776 pKa = 4.25GNFSSEE782 pKa = 4.14GNPTHH787 pKa = 7.18TYY789 pKa = 7.94NTAGTYY795 pKa = 9.4VVNLEE800 pKa = 3.86ISYY803 pKa = 10.95DD804 pKa = 3.93GITNCSADD812 pKa = 3.52LSKK815 pKa = 11.08QFIVNGEE822 pKa = 4.2LNLTLNSTTSSLCEE836 pKa = 3.78GDD838 pKa = 4.01SAVLSLDD845 pKa = 3.3NTYY848 pKa = 11.09EE849 pKa = 4.03SYY851 pKa = 11.23AWDD854 pKa = 3.57TGEE857 pKa = 4.07TTPTITVNEE866 pKa = 4.39GGTYY870 pKa = 9.59SVSVVDD876 pKa = 4.36EE877 pKa = 4.44NGCEE881 pKa = 4.04GSSEE885 pKa = 4.02ISIQTLPIPDD895 pKa = 3.72VSLDD899 pKa = 3.6ASSTAINAGDD909 pKa = 4.03TVTLSASGLLDD920 pKa = 4.07YY921 pKa = 10.58IWYY924 pKa = 10.18ADD926 pKa = 3.6STEE929 pKa = 5.2LEE931 pKa = 4.3LTDD934 pKa = 5.12DD935 pKa = 4.13QIEE938 pKa = 4.22YY939 pKa = 10.94APTNTTTIRR948 pKa = 11.84VEE950 pKa = 4.1GQDD953 pKa = 3.38EE954 pKa = 4.34NGCFGSAEE962 pKa = 3.62ILLNVEE968 pKa = 3.98EE969 pKa = 4.46TNIGDD974 pKa = 4.27RR975 pKa = 11.84IEE977 pKa = 4.05PMKK980 pKa = 10.69FFSPNGDD987 pKa = 3.92AIAQFWRR994 pKa = 11.84IEE996 pKa = 4.14NIEE999 pKa = 5.04DD1000 pKa = 3.38LTQCAVEE1007 pKa = 4.91IYY1009 pKa = 10.28DD1010 pKa = 3.66KK1011 pKa = 11.09QGNKK1015 pKa = 9.38ILEE1018 pKa = 4.35AKK1020 pKa = 9.8PYY1022 pKa = 10.14NNDD1025 pKa = 2.65WDD1027 pKa = 4.0GQINGRR1033 pKa = 11.84PVPDD1037 pKa = 3.24GVYY1040 pKa = 10.31YY1041 pKa = 9.72YY1042 pKa = 10.42VIRR1045 pKa = 11.84CDD1047 pKa = 3.13DD1048 pKa = 3.53TGIVKK1053 pKa = 10.42SGSITLLRR1061 pKa = 4.59
MM1 pKa = 7.53KK2 pKa = 10.38FLPTYY7 pKa = 10.45LFFIGLVFFANQSFGQTSYY26 pKa = 11.07IEE28 pKa = 4.32SVNPKK33 pKa = 9.77NGYY36 pKa = 8.95SGQIINISGVGLAGADD52 pKa = 3.13KK53 pKa = 11.08VFFGSVEE60 pKa = 3.88GKK62 pKa = 10.0IINVEE67 pKa = 3.99DD68 pKa = 3.6QLIEE72 pKa = 4.28AEE74 pKa = 4.58VPAGTTYY81 pKa = 11.68DD82 pKa = 3.77NITVLKK88 pKa = 10.31SSTRR92 pKa = 11.84LSYY95 pKa = 10.85SGEE98 pKa = 4.13HH99 pKa = 6.12FLLSYY104 pKa = 10.67GGEE107 pKa = 4.17QGVSSSDD114 pKa = 3.29FDD116 pKa = 4.11DD117 pKa = 3.97QVDD120 pKa = 3.7VFADD124 pKa = 3.64DD125 pKa = 3.7GLYY128 pKa = 10.86DD129 pKa = 3.68VTLSDD134 pKa = 5.57LDD136 pKa = 4.01GDD138 pKa = 4.25GKK140 pKa = 10.82NDD142 pKa = 3.79IIGANSKK149 pKa = 10.78SNSATILRR157 pKa = 11.84NLSTPSNLSFDD168 pKa = 3.61QRR170 pKa = 11.84NIGIGAPTLNITAGDD185 pKa = 3.97LNGDD189 pKa = 4.23GKK191 pKa = 10.17PDD193 pKa = 3.54VVFSEE198 pKa = 4.79AGDD201 pKa = 3.8GNRR204 pKa = 11.84LIILGNNSSPGSLNFSIQTITLEE227 pKa = 4.3GSSTKK232 pKa = 10.37RR233 pKa = 11.84VVIKK237 pKa = 10.85DD238 pKa = 3.6LDD240 pKa = 4.23LDD242 pKa = 4.09GKK244 pKa = 9.79PDD246 pKa = 3.94LVVSDD251 pKa = 3.65QVNNRR256 pKa = 11.84ILIIKK261 pKa = 7.01NTSSAGTLSFSPEE274 pKa = 3.87IIEE277 pKa = 4.14LTVEE281 pKa = 4.28NAVSTAGLDD290 pKa = 3.76VEE292 pKa = 5.29DD293 pKa = 5.24LNGDD297 pKa = 4.19GKK299 pKa = 10.18PEE301 pKa = 4.73IITHH305 pKa = 5.75QFQTDD310 pKa = 2.85AGGFFIATNQSSPGNFSFTDD330 pKa = 3.69FNKK333 pKa = 10.26YY334 pKa = 7.6STPGTLINLKK344 pKa = 10.6VGDD347 pKa = 4.19INQDD351 pKa = 3.12NKK353 pKa = 10.79PDD355 pKa = 3.4IVATLFLSSSVAVFNNEE372 pKa = 3.7TTDD375 pKa = 3.42TGNVPQFGSAQNLATDD391 pKa = 3.65VRR393 pKa = 11.84PWGLDD398 pKa = 3.33FGDD401 pKa = 4.07MDD403 pKa = 4.88GDD405 pKa = 4.28GIKK408 pKa = 10.45DD409 pKa = 3.22IVVATVGTDD418 pKa = 3.1KK419 pKa = 11.0TINILNNDD427 pKa = 3.19GTGGLNYY434 pKa = 10.06SKK436 pKa = 11.06VSIPVTYY443 pKa = 10.11INRR446 pKa = 11.84NIKK449 pKa = 10.23IGDD452 pKa = 3.35IDD454 pKa = 4.58GDD456 pKa = 4.19SKK458 pKa = 11.36PDD460 pKa = 3.31IVFTSVDD467 pKa = 3.75DD468 pKa = 4.78DD469 pKa = 4.34NNQITASNISILRR482 pKa = 11.84NNQCIIPVITPEE494 pKa = 4.25GPINACEE501 pKa = 4.0GNPVRR506 pKa = 11.84LEE508 pKa = 4.0TQNIEE513 pKa = 3.89GLTFEE518 pKa = 4.78WFQDD522 pKa = 3.41GTSVKK527 pKa = 9.84TGTEE531 pKa = 3.77NFIEE535 pKa = 4.47LNDD538 pKa = 3.57VSATGSYY545 pKa = 8.74TVSIISDD552 pKa = 3.37GGSCQEE558 pKa = 3.35ISEE561 pKa = 4.69AIEE564 pKa = 3.75VNIISEE570 pKa = 4.33GALPSATISSNDD582 pKa = 4.03PICNGGILTLNSSDD596 pKa = 4.89VGATDD601 pKa = 3.64YY602 pKa = 11.03KK603 pKa = 10.17WRR605 pKa = 11.84GPQGYY610 pKa = 6.59TAEE613 pKa = 5.22GITVEE618 pKa = 4.47VDD620 pKa = 3.45DD621 pKa = 5.51FDD623 pKa = 3.89VDD625 pKa = 3.36KK626 pKa = 11.32AGRR629 pKa = 11.84YY630 pKa = 8.86YY631 pKa = 11.22LDD633 pKa = 4.21VYY635 pKa = 11.35SGDD638 pKa = 4.55CIIEE642 pKa = 4.13TTSIVVEE649 pKa = 4.8VIPSPNFSVEE659 pKa = 3.65QSGAGTYY666 pKa = 10.59CEE668 pKa = 4.71GEE670 pKa = 4.36SVTLNVSPSEE680 pKa = 4.06NGFGFQWYY688 pKa = 9.33RR689 pKa = 11.84GTSTISGATSATYY702 pKa = 10.97NPTTDD707 pKa = 2.54GDD709 pKa = 4.64YY710 pKa = 10.97YY711 pKa = 11.65VEE713 pKa = 4.42ITDD716 pKa = 4.58QVNTGCPKK724 pKa = 10.09IYY726 pKa = 10.44SDD728 pKa = 3.67TLQVAFLEE736 pKa = 4.7TPEE739 pKa = 4.31VYY741 pKa = 10.74YY742 pKa = 10.26DD743 pKa = 4.22LPSSACVGIPVSFTNDD759 pKa = 2.5AVVADD764 pKa = 4.67EE765 pKa = 5.0SLAEE769 pKa = 3.94YY770 pKa = 10.2RR771 pKa = 11.84WDD773 pKa = 3.83FGDD776 pKa = 4.25GNFSSEE782 pKa = 4.14GNPTHH787 pKa = 7.18TYY789 pKa = 7.94NTAGTYY795 pKa = 9.4VVNLEE800 pKa = 3.86ISYY803 pKa = 10.95DD804 pKa = 3.93GITNCSADD812 pKa = 3.52LSKK815 pKa = 11.08QFIVNGEE822 pKa = 4.2LNLTLNSTTSSLCEE836 pKa = 3.78GDD838 pKa = 4.01SAVLSLDD845 pKa = 3.3NTYY848 pKa = 11.09EE849 pKa = 4.03SYY851 pKa = 11.23AWDD854 pKa = 3.57TGEE857 pKa = 4.07TTPTITVNEE866 pKa = 4.39GGTYY870 pKa = 9.59SVSVVDD876 pKa = 4.36EE877 pKa = 4.44NGCEE881 pKa = 4.04GSSEE885 pKa = 4.02ISIQTLPIPDD895 pKa = 3.72VSLDD899 pKa = 3.6ASSTAINAGDD909 pKa = 4.03TVTLSASGLLDD920 pKa = 4.07YY921 pKa = 10.58IWYY924 pKa = 10.18ADD926 pKa = 3.6STEE929 pKa = 5.2LEE931 pKa = 4.3LTDD934 pKa = 5.12DD935 pKa = 4.13QIEE938 pKa = 4.22YY939 pKa = 10.94APTNTTTIRR948 pKa = 11.84VEE950 pKa = 4.1GQDD953 pKa = 3.38EE954 pKa = 4.34NGCFGSAEE962 pKa = 3.62ILLNVEE968 pKa = 3.98EE969 pKa = 4.46TNIGDD974 pKa = 4.27RR975 pKa = 11.84IEE977 pKa = 4.05PMKK980 pKa = 10.69FFSPNGDD987 pKa = 3.92AIAQFWRR994 pKa = 11.84IEE996 pKa = 4.14NIEE999 pKa = 5.04DD1000 pKa = 3.38LTQCAVEE1007 pKa = 4.91IYY1009 pKa = 10.28DD1010 pKa = 3.66KK1011 pKa = 11.09QGNKK1015 pKa = 9.38ILEE1018 pKa = 4.35AKK1020 pKa = 9.8PYY1022 pKa = 10.14NNDD1025 pKa = 2.65WDD1027 pKa = 4.0GQINGRR1033 pKa = 11.84PVPDD1037 pKa = 3.24GVYY1040 pKa = 10.31YY1041 pKa = 9.72YY1042 pKa = 10.42VIRR1045 pKa = 11.84CDD1047 pKa = 3.13DD1048 pKa = 3.53TGIVKK1053 pKa = 10.42SGSITLLRR1061 pKa = 4.59
Molecular weight: 113.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4TV29|E4TV29_MARTH Uncharacterized protein OS=Marivirga tractuosa (strain ATCC 23168 / DSM 4126 / NBRC 15989 / NCIMB 1408 / VKM B-1430 / H-43) OX=643867 GN=Ftrac_2140 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 10.52FIIRR6 pKa = 11.84TIIIVVAAHH15 pKa = 6.51FSLLYY20 pKa = 9.97FPWWSMAICAFFAGAIIAGKK40 pKa = 9.73NLSTFFSGFVGLGFLWLIQAFIIHH64 pKa = 6.68NDD66 pKa = 3.01SGGILSNKK74 pKa = 8.65IAEE77 pKa = 4.45LFGLSDD83 pKa = 4.71GIYY86 pKa = 8.41MVVISGIVAALVGGFSTLSGFRR108 pKa = 11.84FRR110 pKa = 11.84KK111 pKa = 8.33MFSRR115 pKa = 11.84NKK117 pKa = 10.28SRR119 pKa = 11.84GLYY122 pKa = 9.44RR123 pKa = 4.85
MM1 pKa = 7.41KK2 pKa = 10.52FIIRR6 pKa = 11.84TIIIVVAAHH15 pKa = 6.51FSLLYY20 pKa = 9.97FPWWSMAICAFFAGAIIAGKK40 pKa = 9.73NLSTFFSGFVGLGFLWLIQAFIIHH64 pKa = 6.68NDD66 pKa = 3.01SGGILSNKK74 pKa = 8.65IAEE77 pKa = 4.45LFGLSDD83 pKa = 4.71GIYY86 pKa = 8.41MVVISGIVAALVGGFSTLSGFRR108 pKa = 11.84FRR110 pKa = 11.84KK111 pKa = 8.33MFSRR115 pKa = 11.84NKK117 pKa = 10.28SRR119 pKa = 11.84GLYY122 pKa = 9.44RR123 pKa = 4.85
Molecular weight: 13.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1335281 |
30 |
3888 |
356.3 |
40.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.343 ± 0.037 | 0.667 ± 0.013 |
5.709 ± 0.034 | 7.416 ± 0.046 |
5.29 ± 0.033 | 6.417 ± 0.043 |
1.738 ± 0.023 | 7.866 ± 0.039 |
7.421 ± 0.07 | 9.392 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.025 | 6.008 ± 0.046 |
3.362 ± 0.021 | 3.672 ± 0.025 |
3.474 ± 0.025 | 6.862 ± 0.044 |
5.067 ± 0.058 | 5.914 ± 0.031 |
1.081 ± 0.014 | 4.035 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
