
Ctenophore-associated circular virus 4
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.18
Get precalculated fractions of proteins
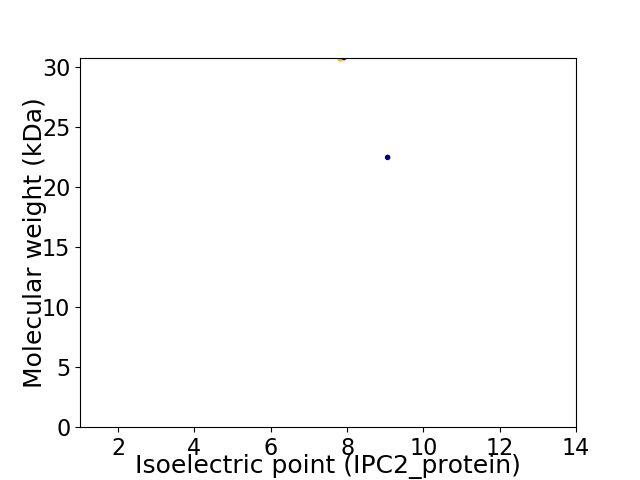
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
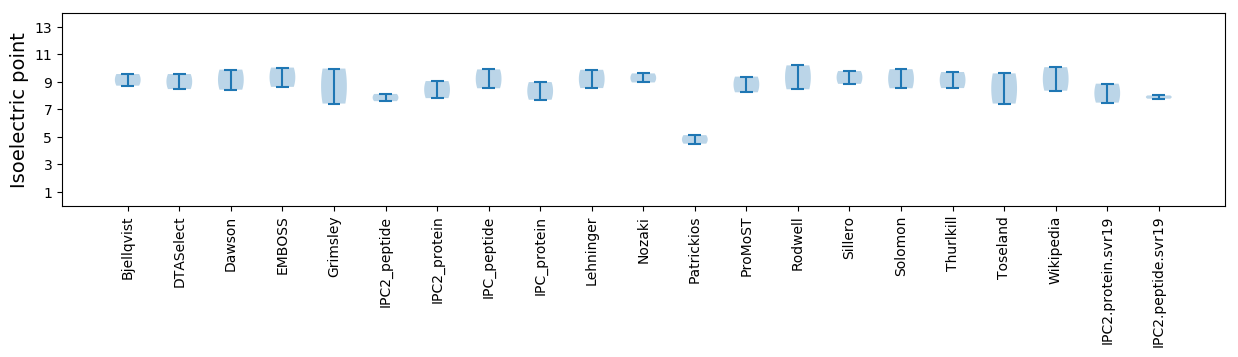
Protein with the lowest isoelectric point:
>tr|A0A141MJB2|A0A141MJB2_9VIRU Coat protein OS=Ctenophore-associated circular virus 4 OX=1778561 PE=4 SV=1
MM1 pKa = 7.64TEE3 pKa = 3.9PTQQDD8 pKa = 3.41TAGNSGGNTKK18 pKa = 10.13GKK20 pKa = 9.69RR21 pKa = 11.84RR22 pKa = 11.84CFAFTWNNYY31 pKa = 8.37PEE33 pKa = 4.13EE34 pKa = 4.21HH35 pKa = 6.57FKK37 pKa = 11.4LFDD40 pKa = 3.48TVAQLSGCKK49 pKa = 9.07FVYY52 pKa = 10.26QEE54 pKa = 3.97EE55 pKa = 4.33TGEE58 pKa = 4.11NGTKK62 pKa = 10.13HH63 pKa = 5.93LQGALYY69 pKa = 10.32FEE71 pKa = 5.2NARR74 pKa = 11.84SWTSVRR80 pKa = 11.84KK81 pKa = 10.14LMDD84 pKa = 3.32GWHH87 pKa = 6.65LEE89 pKa = 4.12VARR92 pKa = 11.84NWMACVKK99 pKa = 10.13YY100 pKa = 9.05ATKK103 pKa = 10.78VEE105 pKa = 4.21TRR107 pKa = 11.84TGDD110 pKa = 3.11QYY112 pKa = 11.96SNFLEE117 pKa = 4.62LRR119 pKa = 11.84PKK121 pKa = 10.75DD122 pKa = 3.61PLEE125 pKa = 4.34GKK127 pKa = 9.42VLHH130 pKa = 6.66PWQSDD135 pKa = 3.53VMNLISSDD143 pKa = 3.61CSNDD147 pKa = 3.25RR148 pKa = 11.84TVHH151 pKa = 5.4WFVDD155 pKa = 3.69SEE157 pKa = 4.46GAKK160 pKa = 10.6GKK162 pKa = 8.94TSLCKK167 pKa = 10.29HH168 pKa = 5.7ILLNKK173 pKa = 9.59VGWLFISAGKK183 pKa = 10.6GGDD186 pKa = 3.32ILCAVARR193 pKa = 11.84YY194 pKa = 8.82LKK196 pKa = 10.45SGKK199 pKa = 9.53KK200 pKa = 9.77LRR202 pKa = 11.84GVLANYY208 pKa = 10.32SRR210 pKa = 11.84DD211 pKa = 3.42KK212 pKa = 11.15EE213 pKa = 4.41GFISYY218 pKa = 10.21SALEE222 pKa = 4.22SIKK225 pKa = 10.96DD226 pKa = 3.53GLIFSTKK233 pKa = 10.14YY234 pKa = 10.04EE235 pKa = 4.15SQHH238 pKa = 7.27AIFDD242 pKa = 4.19SPHH245 pKa = 6.18VICFANWIPDD255 pKa = 3.39KK256 pKa = 11.07RR257 pKa = 11.84KK258 pKa = 10.36LSDD261 pKa = 3.73DD262 pKa = 3.12RR263 pKa = 11.84WDD265 pKa = 3.2IRR267 pKa = 11.84TLL269 pKa = 3.45
MM1 pKa = 7.64TEE3 pKa = 3.9PTQQDD8 pKa = 3.41TAGNSGGNTKK18 pKa = 10.13GKK20 pKa = 9.69RR21 pKa = 11.84RR22 pKa = 11.84CFAFTWNNYY31 pKa = 8.37PEE33 pKa = 4.13EE34 pKa = 4.21HH35 pKa = 6.57FKK37 pKa = 11.4LFDD40 pKa = 3.48TVAQLSGCKK49 pKa = 9.07FVYY52 pKa = 10.26QEE54 pKa = 3.97EE55 pKa = 4.33TGEE58 pKa = 4.11NGTKK62 pKa = 10.13HH63 pKa = 5.93LQGALYY69 pKa = 10.32FEE71 pKa = 5.2NARR74 pKa = 11.84SWTSVRR80 pKa = 11.84KK81 pKa = 10.14LMDD84 pKa = 3.32GWHH87 pKa = 6.65LEE89 pKa = 4.12VARR92 pKa = 11.84NWMACVKK99 pKa = 10.13YY100 pKa = 9.05ATKK103 pKa = 10.78VEE105 pKa = 4.21TRR107 pKa = 11.84TGDD110 pKa = 3.11QYY112 pKa = 11.96SNFLEE117 pKa = 4.62LRR119 pKa = 11.84PKK121 pKa = 10.75DD122 pKa = 3.61PLEE125 pKa = 4.34GKK127 pKa = 9.42VLHH130 pKa = 6.66PWQSDD135 pKa = 3.53VMNLISSDD143 pKa = 3.61CSNDD147 pKa = 3.25RR148 pKa = 11.84TVHH151 pKa = 5.4WFVDD155 pKa = 3.69SEE157 pKa = 4.46GAKK160 pKa = 10.6GKK162 pKa = 8.94TSLCKK167 pKa = 10.29HH168 pKa = 5.7ILLNKK173 pKa = 9.59VGWLFISAGKK183 pKa = 10.6GGDD186 pKa = 3.32ILCAVARR193 pKa = 11.84YY194 pKa = 8.82LKK196 pKa = 10.45SGKK199 pKa = 9.53KK200 pKa = 9.77LRR202 pKa = 11.84GVLANYY208 pKa = 10.32SRR210 pKa = 11.84DD211 pKa = 3.42KK212 pKa = 11.15EE213 pKa = 4.41GFISYY218 pKa = 10.21SALEE222 pKa = 4.22SIKK225 pKa = 10.96DD226 pKa = 3.53GLIFSTKK233 pKa = 10.14YY234 pKa = 10.04EE235 pKa = 4.15SQHH238 pKa = 7.27AIFDD242 pKa = 4.19SPHH245 pKa = 6.18VICFANWIPDD255 pKa = 3.39KK256 pKa = 11.07RR257 pKa = 11.84KK258 pKa = 10.36LSDD261 pKa = 3.73DD262 pKa = 3.12RR263 pKa = 11.84WDD265 pKa = 3.2IRR267 pKa = 11.84TLL269 pKa = 3.45
Molecular weight: 30.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJB2|A0A141MJB2_9VIRU Coat protein OS=Ctenophore-associated circular virus 4 OX=1778561 PE=4 SV=1
MM1 pKa = 7.12VRR3 pKa = 11.84KK4 pKa = 7.74TKK6 pKa = 10.62KK7 pKa = 9.79KK8 pKa = 10.25GKK10 pKa = 9.46QLVTKK15 pKa = 10.25QYY17 pKa = 11.68LNAKK21 pKa = 8.55LNKK24 pKa = 10.14AIEE27 pKa = 4.12TKK29 pKa = 10.64YY30 pKa = 10.84YY31 pKa = 7.89YY32 pKa = 10.14TLGASSPDD40 pKa = 3.35TGGSLVTVSNGIAQGDD56 pKa = 4.22AYY58 pKa = 10.97NEE60 pKa = 4.01RR61 pKa = 11.84LGDD64 pKa = 4.38RR65 pKa = 11.84ITPIHH70 pKa = 6.03CHH72 pKa = 5.49IKK74 pKa = 9.65GTVTLADD81 pKa = 3.28TTNWVRR87 pKa = 11.84IVLIRR92 pKa = 11.84WKK94 pKa = 10.17QDD96 pKa = 3.18YY97 pKa = 11.46GDD99 pKa = 4.27LTDD102 pKa = 4.3ANQIFEE108 pKa = 4.46ATGATTSPYY117 pKa = 10.58SLFVKK122 pKa = 10.44EE123 pKa = 4.02PDD125 pKa = 2.87NRR127 pKa = 11.84ARR129 pKa = 11.84FDD131 pKa = 3.49VLADD135 pKa = 3.62RR136 pKa = 11.84LFNLHH141 pKa = 6.65SNEE144 pKa = 3.89PQKK147 pKa = 11.38AFDD150 pKa = 3.59IRR152 pKa = 11.84VSSRR156 pKa = 11.84KK157 pKa = 8.87LHH159 pKa = 5.58SRR161 pKa = 11.84PVSFTAGGSGGNGQLVMVTISDD183 pKa = 4.01SAAVAHH189 pKa = 6.29PSISYY194 pKa = 11.12YY195 pKa = 11.5LMLKK199 pKa = 10.48YY200 pKa = 10.34KK201 pKa = 10.57DD202 pKa = 3.42AA203 pKa = 5.65
MM1 pKa = 7.12VRR3 pKa = 11.84KK4 pKa = 7.74TKK6 pKa = 10.62KK7 pKa = 9.79KK8 pKa = 10.25GKK10 pKa = 9.46QLVTKK15 pKa = 10.25QYY17 pKa = 11.68LNAKK21 pKa = 8.55LNKK24 pKa = 10.14AIEE27 pKa = 4.12TKK29 pKa = 10.64YY30 pKa = 10.84YY31 pKa = 7.89YY32 pKa = 10.14TLGASSPDD40 pKa = 3.35TGGSLVTVSNGIAQGDD56 pKa = 4.22AYY58 pKa = 10.97NEE60 pKa = 4.01RR61 pKa = 11.84LGDD64 pKa = 4.38RR65 pKa = 11.84ITPIHH70 pKa = 6.03CHH72 pKa = 5.49IKK74 pKa = 9.65GTVTLADD81 pKa = 3.28TTNWVRR87 pKa = 11.84IVLIRR92 pKa = 11.84WKK94 pKa = 10.17QDD96 pKa = 3.18YY97 pKa = 11.46GDD99 pKa = 4.27LTDD102 pKa = 4.3ANQIFEE108 pKa = 4.46ATGATTSPYY117 pKa = 10.58SLFVKK122 pKa = 10.44EE123 pKa = 4.02PDD125 pKa = 2.87NRR127 pKa = 11.84ARR129 pKa = 11.84FDD131 pKa = 3.49VLADD135 pKa = 3.62RR136 pKa = 11.84LFNLHH141 pKa = 6.65SNEE144 pKa = 3.89PQKK147 pKa = 11.38AFDD150 pKa = 3.59IRR152 pKa = 11.84VSSRR156 pKa = 11.84KK157 pKa = 8.87LHH159 pKa = 5.58SRR161 pKa = 11.84PVSFTAGGSGGNGQLVMVTISDD183 pKa = 4.01SAAVAHH189 pKa = 6.29PSISYY194 pKa = 11.12YY195 pKa = 11.5LMLKK199 pKa = 10.48YY200 pKa = 10.34KK201 pKa = 10.57DD202 pKa = 3.42AA203 pKa = 5.65
Molecular weight: 22.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
472 |
203 |
269 |
236.0 |
26.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.992 ± 0.834 | 1.695 ± 0.725 |
6.356 ± 0.029 | 4.237 ± 1.07 |
4.025 ± 0.645 | 7.627 ± 0.144 |
2.754 ± 0.176 | 4.661 ± 0.457 |
8.263 ± 0.23 | 8.475 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.483 ± 0.003 | 4.873 ± 0.032 |
2.966 ± 0.291 | 3.178 ± 0.163 |
5.297 ± 0.074 | 7.839 ± 0.026 |
6.992 ± 0.834 | 5.932 ± 0.582 |
2.331 ± 0.811 | 4.025 ± 0.543 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
