
Oceanithermus profundus (strain DSM 14977 / NBRC 100410 / VKM B-2274 / 506)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Thermales; Thermaceae; Oceanithermus; Oceanithermus profundus
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
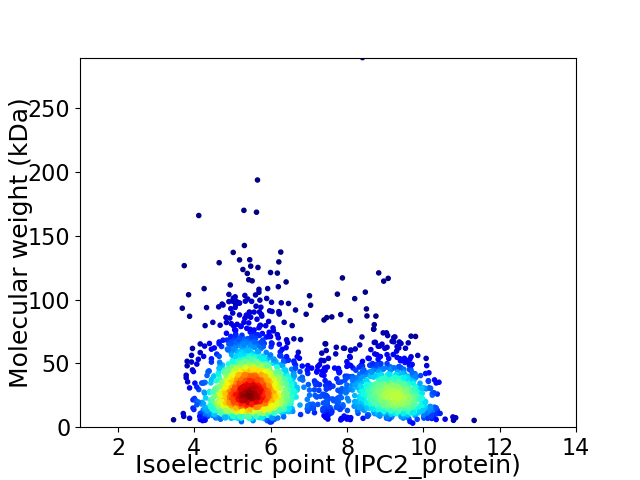
Virtual 2D-PAGE plot for 2372 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4UA54|E4UA54_OCEP5 Small multidrug resistance protein OS=Oceanithermus profundus (strain DSM 14977 / NBRC 100410 / VKM B-2274 / 506) OX=670487 GN=Ocepr_1980 PE=3 SV=1
MM1 pKa = 7.2LTHH4 pKa = 6.37LRR6 pKa = 11.84YY7 pKa = 9.35RR8 pKa = 11.84VRR10 pKa = 11.84FAAFVLGVMGLLAACSQPAGGAGGFSLSLEE40 pKa = 4.29PTSVTVAAGGSTTVRR55 pKa = 11.84LDD57 pKa = 3.17VGSEE61 pKa = 4.02GGFSGEE67 pKa = 3.72VSLALEE73 pKa = 4.49GLAGATVSPARR84 pKa = 11.84VQVPGGPYY92 pKa = 9.73TLTLAVDD99 pKa = 4.05DD100 pKa = 4.59SVAPGSYY107 pKa = 10.88DD108 pKa = 3.28LALVGASGSLMGRR121 pKa = 11.84ATFTLAVTDD130 pKa = 3.75ATPDD134 pKa = 3.41FEE136 pKa = 4.93LALSAVDD143 pKa = 4.48LAADD147 pKa = 3.96PGGSVGTALTVTPSGGFAGTLTLSLVDD174 pKa = 4.99ASDD177 pKa = 3.7APAAGFTLVPASLYY191 pKa = 10.73VSGIVQQPLSIQVAGDD207 pKa = 3.72VGPGTYY213 pKa = 10.18PLRR216 pKa = 11.84LKK218 pKa = 9.66VTGDD222 pKa = 3.52GVTHH226 pKa = 6.02YY227 pKa = 11.21ADD229 pKa = 3.69LAVTVSGLEE238 pKa = 3.83LALTSTEE245 pKa = 4.31LYY247 pKa = 10.5APQGGTATATLLVNAVGVSGDD268 pKa = 3.64LALALASADD277 pKa = 3.74GGPAPTGLALQPSSVAVPGGPYY299 pKa = 10.55ALSLSVDD306 pKa = 3.27AGVATGSYY314 pKa = 11.07DD315 pKa = 4.0LLLKK319 pKa = 9.68GTLGSFEE326 pKa = 4.43RR327 pKa = 11.84SVAFTLTVQPPPEE340 pKa = 4.24FTTSVSPDD348 pKa = 3.21ALTVEE353 pKa = 4.72VGSSGTLYY361 pKa = 11.19LEE363 pKa = 4.51LTFQADD369 pKa = 3.7FNGDD373 pKa = 2.69IGFALEE379 pKa = 4.41LADD382 pKa = 5.49GSAAPAGLALDD393 pKa = 3.82PTVVSMAANSGDD405 pKa = 3.66TNYY408 pKa = 10.38VALDD412 pKa = 3.67LAVADD417 pKa = 4.13TMAPGTYY424 pKa = 10.13ALRR427 pKa = 11.84VRR429 pKa = 11.84ASSSNTTQYY438 pKa = 11.73AGFSLTVPVPPDD450 pKa = 3.31FTLSLDD456 pKa = 3.39PTIADD461 pKa = 3.56VVAGGSTTVRR471 pKa = 11.84LDD473 pKa = 3.58AVPHH477 pKa = 6.02NGFAGVVQLSLVDD490 pKa = 4.01SSNGAAPSGMTLEE503 pKa = 4.36PTSVDD508 pKa = 3.37LSGGAVSQTLTLRR521 pKa = 11.84VDD523 pKa = 3.25EE524 pKa = 4.78GVLVDD529 pKa = 5.05RR530 pKa = 11.84YY531 pKa = 9.6HH532 pKa = 6.16LQVVGSGGGTTQTANLSLSVQDD554 pKa = 4.7FSLSFGTTTPSLAADD569 pKa = 3.53QGGSATTTLTVQVSQPDD586 pKa = 4.08TYY588 pKa = 10.81STFPGPVALEE598 pKa = 4.48LATQDD603 pKa = 4.55GSPLPPGLTLSPVSVDD619 pKa = 3.55LSSGYY624 pKa = 11.13VNVNLTVTADD634 pKa = 3.09ASTPPGSYY642 pKa = 10.67ALMVIGRR649 pKa = 11.84AGGTARR655 pKa = 11.84AAAFGLNVRR664 pKa = 11.84GFDD667 pKa = 3.56VALGASEE674 pKa = 4.06LAFWTEE680 pKa = 3.9GSGDD684 pKa = 3.9LGLTLTPGGGFDD696 pKa = 3.76GTVSLSLEE704 pKa = 4.29AQDD707 pKa = 5.47GSAAPAGLTLSPSSVSVSSTTSVTLSVAADD737 pKa = 3.39ASVAAGVYY745 pKa = 9.13PLRR748 pKa = 11.84LKK750 pKa = 10.6AVSGSIVHH758 pKa = 6.16YY759 pKa = 11.12ADD761 pKa = 3.45FTLKK765 pKa = 10.95VGDD768 pKa = 4.21FDD770 pKa = 5.0LALDD774 pKa = 4.22ASSLAVWQNDD784 pKa = 2.89RR785 pKa = 11.84SGFGVTVTPSGGFAGTVNLYY805 pKa = 9.7LAPQVGATGPTGVSLDD821 pKa = 3.78PSALDD826 pKa = 3.27VSGGVSQTVNVVAADD841 pKa = 3.6AASPGSFAMQLEE853 pKa = 3.89ARR855 pKa = 11.84AYY857 pKa = 9.07LGGVARR863 pKa = 11.84SRR865 pKa = 11.84TVPFTLEE872 pKa = 3.83VKK874 pKa = 10.56GFLVEE879 pKa = 4.56LSEE882 pKa = 5.08DD883 pKa = 3.58NYY885 pKa = 11.06FEE887 pKa = 4.63ARR889 pKa = 11.84GGSASGQLTLTAYY902 pKa = 10.29GLNDD906 pKa = 3.6VVALALLDD914 pKa = 3.93PSGAAPAGIALAPASVAASDD934 pKa = 3.87GAGTHH939 pKa = 7.18TITIGTDD946 pKa = 2.82ANVDD950 pKa = 3.36YY951 pKa = 11.17DD952 pKa = 3.73RR953 pKa = 11.84TYY955 pKa = 11.72GLDD958 pKa = 4.65LVATWGSLEE967 pKa = 3.91RR968 pKa = 11.84RR969 pKa = 11.84LAINLTVYY977 pKa = 10.86RR978 pKa = 11.84LTEE981 pKa = 3.77YY982 pKa = 9.58WVPRR986 pKa = 11.84DD987 pKa = 3.56PGTDD991 pKa = 3.2YY992 pKa = 11.48DD993 pKa = 3.99LWGVAYY999 pKa = 10.41GEE1001 pKa = 4.57VNGSGLFVAVGGGQLSLNGSSYY1023 pKa = 11.0GVAATSTDD1031 pKa = 3.4GASWSLTEE1039 pKa = 5.83DD1040 pKa = 3.0ISSNPLLGIGYY1051 pKa = 10.24GGGTFVAVGRR1061 pKa = 11.84ACEE1064 pKa = 3.72IVTYY1068 pKa = 10.57DD1069 pKa = 3.21GTGWTTRR1076 pKa = 11.84ANPDD1080 pKa = 3.54AYY1082 pKa = 10.89CSANLNDD1089 pKa = 3.49VAYY1092 pKa = 10.68SGTRR1096 pKa = 11.84FVAVGDD1102 pKa = 3.85SGTVLWSNDD1111 pKa = 2.92GASWSSATSGVSGDD1125 pKa = 4.63LYY1127 pKa = 11.14AITYY1131 pKa = 10.39GNGTFVAVGAGGTVLTSTDD1150 pKa = 3.38GLSWSAQASGTTADD1164 pKa = 4.32LSGIAYY1170 pKa = 10.33GDD1172 pKa = 3.61GTFVAVGAGGTVLTSTDD1189 pKa = 3.33GASWSLGDD1197 pKa = 5.07LGTSGDD1203 pKa = 3.94ATGVAYY1209 pKa = 9.8GYY1211 pKa = 10.82DD1212 pKa = 3.53WDD1214 pKa = 4.06NVGVFVVTTNEE1225 pKa = 4.18PNTGIHH1231 pKa = 6.03TSSDD1235 pKa = 3.4AGSSWSSQSSGGLGDD1250 pKa = 3.9ALEE1253 pKa = 4.16AAGYY1257 pKa = 8.59GDD1259 pKa = 3.54HH1260 pKa = 6.48VFVVVGQLGALGTSPP1275 pKa = 4.6
MM1 pKa = 7.2LTHH4 pKa = 6.37LRR6 pKa = 11.84YY7 pKa = 9.35RR8 pKa = 11.84VRR10 pKa = 11.84FAAFVLGVMGLLAACSQPAGGAGGFSLSLEE40 pKa = 4.29PTSVTVAAGGSTTVRR55 pKa = 11.84LDD57 pKa = 3.17VGSEE61 pKa = 4.02GGFSGEE67 pKa = 3.72VSLALEE73 pKa = 4.49GLAGATVSPARR84 pKa = 11.84VQVPGGPYY92 pKa = 9.73TLTLAVDD99 pKa = 4.05DD100 pKa = 4.59SVAPGSYY107 pKa = 10.88DD108 pKa = 3.28LALVGASGSLMGRR121 pKa = 11.84ATFTLAVTDD130 pKa = 3.75ATPDD134 pKa = 3.41FEE136 pKa = 4.93LALSAVDD143 pKa = 4.48LAADD147 pKa = 3.96PGGSVGTALTVTPSGGFAGTLTLSLVDD174 pKa = 4.99ASDD177 pKa = 3.7APAAGFTLVPASLYY191 pKa = 10.73VSGIVQQPLSIQVAGDD207 pKa = 3.72VGPGTYY213 pKa = 10.18PLRR216 pKa = 11.84LKK218 pKa = 9.66VTGDD222 pKa = 3.52GVTHH226 pKa = 6.02YY227 pKa = 11.21ADD229 pKa = 3.69LAVTVSGLEE238 pKa = 3.83LALTSTEE245 pKa = 4.31LYY247 pKa = 10.5APQGGTATATLLVNAVGVSGDD268 pKa = 3.64LALALASADD277 pKa = 3.74GGPAPTGLALQPSSVAVPGGPYY299 pKa = 10.55ALSLSVDD306 pKa = 3.27AGVATGSYY314 pKa = 11.07DD315 pKa = 4.0LLLKK319 pKa = 9.68GTLGSFEE326 pKa = 4.43RR327 pKa = 11.84SVAFTLTVQPPPEE340 pKa = 4.24FTTSVSPDD348 pKa = 3.21ALTVEE353 pKa = 4.72VGSSGTLYY361 pKa = 11.19LEE363 pKa = 4.51LTFQADD369 pKa = 3.7FNGDD373 pKa = 2.69IGFALEE379 pKa = 4.41LADD382 pKa = 5.49GSAAPAGLALDD393 pKa = 3.82PTVVSMAANSGDD405 pKa = 3.66TNYY408 pKa = 10.38VALDD412 pKa = 3.67LAVADD417 pKa = 4.13TMAPGTYY424 pKa = 10.13ALRR427 pKa = 11.84VRR429 pKa = 11.84ASSSNTTQYY438 pKa = 11.73AGFSLTVPVPPDD450 pKa = 3.31FTLSLDD456 pKa = 3.39PTIADD461 pKa = 3.56VVAGGSTTVRR471 pKa = 11.84LDD473 pKa = 3.58AVPHH477 pKa = 6.02NGFAGVVQLSLVDD490 pKa = 4.01SSNGAAPSGMTLEE503 pKa = 4.36PTSVDD508 pKa = 3.37LSGGAVSQTLTLRR521 pKa = 11.84VDD523 pKa = 3.25EE524 pKa = 4.78GVLVDD529 pKa = 5.05RR530 pKa = 11.84YY531 pKa = 9.6HH532 pKa = 6.16LQVVGSGGGTTQTANLSLSVQDD554 pKa = 4.7FSLSFGTTTPSLAADD569 pKa = 3.53QGGSATTTLTVQVSQPDD586 pKa = 4.08TYY588 pKa = 10.81STFPGPVALEE598 pKa = 4.48LATQDD603 pKa = 4.55GSPLPPGLTLSPVSVDD619 pKa = 3.55LSSGYY624 pKa = 11.13VNVNLTVTADD634 pKa = 3.09ASTPPGSYY642 pKa = 10.67ALMVIGRR649 pKa = 11.84AGGTARR655 pKa = 11.84AAAFGLNVRR664 pKa = 11.84GFDD667 pKa = 3.56VALGASEE674 pKa = 4.06LAFWTEE680 pKa = 3.9GSGDD684 pKa = 3.9LGLTLTPGGGFDD696 pKa = 3.76GTVSLSLEE704 pKa = 4.29AQDD707 pKa = 5.47GSAAPAGLTLSPSSVSVSSTTSVTLSVAADD737 pKa = 3.39ASVAAGVYY745 pKa = 9.13PLRR748 pKa = 11.84LKK750 pKa = 10.6AVSGSIVHH758 pKa = 6.16YY759 pKa = 11.12ADD761 pKa = 3.45FTLKK765 pKa = 10.95VGDD768 pKa = 4.21FDD770 pKa = 5.0LALDD774 pKa = 4.22ASSLAVWQNDD784 pKa = 2.89RR785 pKa = 11.84SGFGVTVTPSGGFAGTVNLYY805 pKa = 9.7LAPQVGATGPTGVSLDD821 pKa = 3.78PSALDD826 pKa = 3.27VSGGVSQTVNVVAADD841 pKa = 3.6AASPGSFAMQLEE853 pKa = 3.89ARR855 pKa = 11.84AYY857 pKa = 9.07LGGVARR863 pKa = 11.84SRR865 pKa = 11.84TVPFTLEE872 pKa = 3.83VKK874 pKa = 10.56GFLVEE879 pKa = 4.56LSEE882 pKa = 5.08DD883 pKa = 3.58NYY885 pKa = 11.06FEE887 pKa = 4.63ARR889 pKa = 11.84GGSASGQLTLTAYY902 pKa = 10.29GLNDD906 pKa = 3.6VVALALLDD914 pKa = 3.93PSGAAPAGIALAPASVAASDD934 pKa = 3.87GAGTHH939 pKa = 7.18TITIGTDD946 pKa = 2.82ANVDD950 pKa = 3.36YY951 pKa = 11.17DD952 pKa = 3.73RR953 pKa = 11.84TYY955 pKa = 11.72GLDD958 pKa = 4.65LVATWGSLEE967 pKa = 3.91RR968 pKa = 11.84RR969 pKa = 11.84LAINLTVYY977 pKa = 10.86RR978 pKa = 11.84LTEE981 pKa = 3.77YY982 pKa = 9.58WVPRR986 pKa = 11.84DD987 pKa = 3.56PGTDD991 pKa = 3.2YY992 pKa = 11.48DD993 pKa = 3.99LWGVAYY999 pKa = 10.41GEE1001 pKa = 4.57VNGSGLFVAVGGGQLSLNGSSYY1023 pKa = 11.0GVAATSTDD1031 pKa = 3.4GASWSLTEE1039 pKa = 5.83DD1040 pKa = 3.0ISSNPLLGIGYY1051 pKa = 10.24GGGTFVAVGRR1061 pKa = 11.84ACEE1064 pKa = 3.72IVTYY1068 pKa = 10.57DD1069 pKa = 3.21GTGWTTRR1076 pKa = 11.84ANPDD1080 pKa = 3.54AYY1082 pKa = 10.89CSANLNDD1089 pKa = 3.49VAYY1092 pKa = 10.68SGTRR1096 pKa = 11.84FVAVGDD1102 pKa = 3.85SGTVLWSNDD1111 pKa = 2.92GASWSSATSGVSGDD1125 pKa = 4.63LYY1127 pKa = 11.14AITYY1131 pKa = 10.39GNGTFVAVGAGGTVLTSTDD1150 pKa = 3.38GLSWSAQASGTTADD1164 pKa = 4.32LSGIAYY1170 pKa = 10.33GDD1172 pKa = 3.61GTFVAVGAGGTVLTSTDD1189 pKa = 3.33GASWSLGDD1197 pKa = 5.07LGTSGDD1203 pKa = 3.94ATGVAYY1209 pKa = 9.8GYY1211 pKa = 10.82DD1212 pKa = 3.53WDD1214 pKa = 4.06NVGVFVVTTNEE1225 pKa = 4.18PNTGIHH1231 pKa = 6.03TSSDD1235 pKa = 3.4AGSSWSSQSSGGLGDD1250 pKa = 3.9ALEE1253 pKa = 4.16AAGYY1257 pKa = 8.59GDD1259 pKa = 3.54HH1260 pKa = 6.48VFVVVGQLGALGTSPP1275 pKa = 4.6
Molecular weight: 126.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4U4H4|E4U4H4_OCEP5 Thymidine kinase OS=Oceanithermus profundus (strain DSM 14977 / NBRC 100410 / VKM B-2274 / 506) OX=670487 GN=tdk PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.45RR3 pKa = 11.84TWQPNRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.75RR12 pKa = 11.84AKK14 pKa = 8.56THH16 pKa = 5.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 8.77VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.38KK2 pKa = 9.45RR3 pKa = 11.84TWQPNRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.75RR12 pKa = 11.84AKK14 pKa = 8.56THH16 pKa = 5.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 8.77VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
750473 |
30 |
2686 |
316.4 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.74 ± 0.073 | 0.433 ± 0.015 |
4.711 ± 0.038 | 7.414 ± 0.063 |
3.599 ± 0.032 | 9.096 ± 0.052 |
1.877 ± 0.021 | 3.459 ± 0.043 |
2.928 ± 0.044 | 11.98 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.768 ± 0.021 | 1.943 ± 0.027 |
6.034 ± 0.039 | 2.561 ± 0.025 |
8.171 ± 0.073 | 3.867 ± 0.031 |
4.476 ± 0.041 | 8.387 ± 0.052 |
1.701 ± 0.027 | 2.855 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
