
Mosquito VEM Anellovirus SDBVL B
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
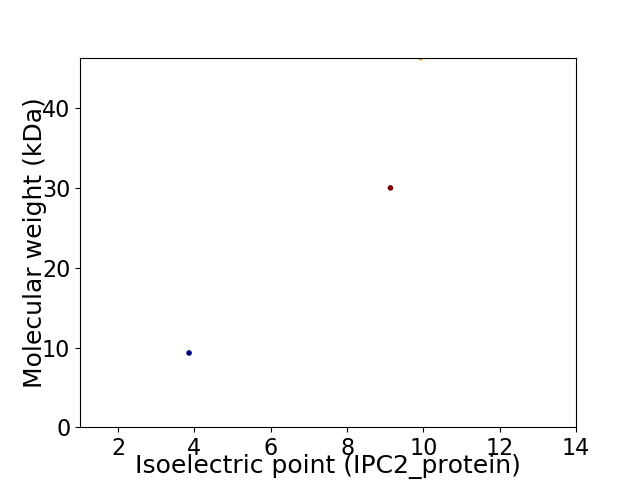
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
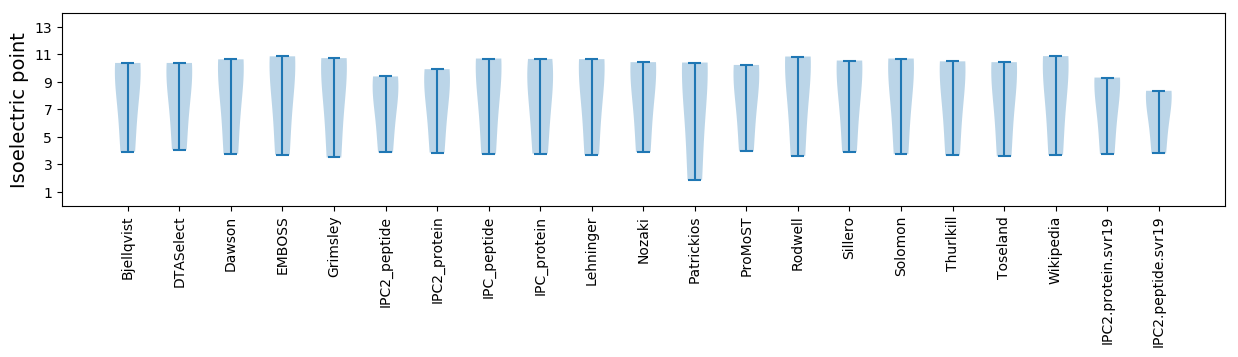
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6KID2|F6KID2_9VIRU Capsid protein OS=Mosquito VEM Anellovirus SDBVL B OX=1034791 PE=3 SV=1
MM1 pKa = 7.75AFTASLGQDD10 pKa = 3.09EE11 pKa = 5.51DD12 pKa = 3.11RR13 pKa = 11.84WKK15 pKa = 11.13LVIQTSHH22 pKa = 6.51MLFCDD27 pKa = 3.24CGDD30 pKa = 3.92YY31 pKa = 11.01VKK33 pKa = 10.6HH34 pKa = 5.89LQNILGPQWLCTSEE48 pKa = 4.76EE49 pKa = 4.07EE50 pKa = 5.34GGTGDD55 pKa = 3.89QSGAGEE61 pKa = 4.25ATGEE65 pKa = 4.22DD66 pKa = 3.86PTDD69 pKa = 3.52QLMAAALDD77 pKa = 3.9AAEE80 pKa = 4.68EE81 pKa = 4.13LAGAPGSGG89 pKa = 3.22
MM1 pKa = 7.75AFTASLGQDD10 pKa = 3.09EE11 pKa = 5.51DD12 pKa = 3.11RR13 pKa = 11.84WKK15 pKa = 11.13LVIQTSHH22 pKa = 6.51MLFCDD27 pKa = 3.24CGDD30 pKa = 3.92YY31 pKa = 11.01VKK33 pKa = 10.6HH34 pKa = 5.89LQNILGPQWLCTSEE48 pKa = 4.76EE49 pKa = 4.07EE50 pKa = 5.34GGTGDD55 pKa = 3.89QSGAGEE61 pKa = 4.25ATGEE65 pKa = 4.22DD66 pKa = 3.86PTDD69 pKa = 3.52QLMAAALDD77 pKa = 3.9AAEE80 pKa = 4.68EE81 pKa = 4.13LAGAPGSGG89 pKa = 3.22
Molecular weight: 9.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6KID3|F6KID3_9VIRU Uncharacterized protein OS=Mosquito VEM Anellovirus SDBVL B OX=1034791 PE=4 SV=1
MM1 pKa = 7.54ALYY4 pKa = 9.76FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 8.08WRR13 pKa = 11.84PKK15 pKa = 6.8WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GYY21 pKa = 8.14WRR23 pKa = 11.84RR24 pKa = 11.84SYY26 pKa = 10.7RR27 pKa = 11.84PAYY30 pKa = 9.45GRR32 pKa = 11.84RR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84VRR49 pKa = 11.84VLTEE53 pKa = 3.47RR54 pKa = 11.84QPRR57 pKa = 11.84YY58 pKa = 8.33WRR60 pKa = 11.84RR61 pKa = 11.84CTIRR65 pKa = 11.84GWWPVLITGRR75 pKa = 11.84KK76 pKa = 7.69YY77 pKa = 10.69PSTAAAGDD85 pKa = 3.76DD86 pKa = 4.37RR87 pKa = 11.84GTQMLTFKK95 pKa = 10.27PFQPFGSKK103 pKa = 10.44LSNAWFYY110 pKa = 11.34RR111 pKa = 11.84DD112 pKa = 3.59YY113 pKa = 11.05EE114 pKa = 4.16IVRR117 pKa = 11.84GGFSAGTFSLQALYY131 pKa = 9.61MEE133 pKa = 5.38HH134 pKa = 6.84LMCRR138 pKa = 11.84NRR140 pKa = 11.84WSQSNCGFDD149 pKa = 3.28LASYY153 pKa = 10.31RR154 pKa = 11.84GTKK157 pKa = 10.09LYY159 pKa = 9.93FVPHH163 pKa = 6.85PNYY166 pKa = 10.77DD167 pKa = 4.12YY168 pKa = 11.1IVLIDD173 pKa = 3.4PEE175 pKa = 4.31YY176 pKa = 11.01RR177 pKa = 11.84KK178 pKa = 10.14FEE180 pKa = 3.91QWIKK184 pKa = 10.76QPMHH188 pKa = 6.55PGVLITHH195 pKa = 6.17PQSRR199 pKa = 11.84IIRR202 pKa = 11.84SITHH206 pKa = 6.37AGPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 8.68MPKK215 pKa = 8.94MFVPPPSTMNSGWQWMGEE233 pKa = 4.03LANEE237 pKa = 4.3GLFAFYY243 pKa = 10.2VQWIDD248 pKa = 5.72LGAPWLGDD256 pKa = 3.3VQNPNQVKK264 pKa = 8.2WWKK267 pKa = 9.23TGDD270 pKa = 3.42ATTKK274 pKa = 10.19PDD276 pKa = 3.25WVTKK280 pKa = 9.9GIEE283 pKa = 4.16MQGKK287 pKa = 6.7TVQEE291 pKa = 4.21GLDD294 pKa = 3.26AYY296 pKa = 8.81YY297 pKa = 9.31TQAGQGGKK305 pKa = 9.51KK306 pKa = 8.22WVNPGGAPYY315 pKa = 10.5QLGFGPFVFKK325 pKa = 10.71GSHH328 pKa = 6.15SGDD331 pKa = 3.41QEE333 pKa = 4.54HH334 pKa = 6.55YY335 pKa = 9.54PQICFFYY342 pKa = 10.47KK343 pKa = 10.46SYY345 pKa = 8.74WQWGGSTTSIKK356 pKa = 10.46NVCDD360 pKa = 3.8PKK362 pKa = 11.2DD363 pKa = 3.79NPNAEE368 pKa = 4.44YY369 pKa = 10.78LSAWNRR375 pKa = 11.84AGFQPAQARR384 pKa = 11.84NNDD387 pKa = 3.88IYY389 pKa = 11.22NQPP392 pKa = 3.25
MM1 pKa = 7.54ALYY4 pKa = 9.76FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 8.08WRR13 pKa = 11.84PKK15 pKa = 6.8WRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GYY21 pKa = 8.14WRR23 pKa = 11.84RR24 pKa = 11.84SYY26 pKa = 10.7RR27 pKa = 11.84PAYY30 pKa = 9.45GRR32 pKa = 11.84RR33 pKa = 11.84LRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84TRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84VRR49 pKa = 11.84VLTEE53 pKa = 3.47RR54 pKa = 11.84QPRR57 pKa = 11.84YY58 pKa = 8.33WRR60 pKa = 11.84RR61 pKa = 11.84CTIRR65 pKa = 11.84GWWPVLITGRR75 pKa = 11.84KK76 pKa = 7.69YY77 pKa = 10.69PSTAAAGDD85 pKa = 3.76DD86 pKa = 4.37RR87 pKa = 11.84GTQMLTFKK95 pKa = 10.27PFQPFGSKK103 pKa = 10.44LSNAWFYY110 pKa = 11.34RR111 pKa = 11.84DD112 pKa = 3.59YY113 pKa = 11.05EE114 pKa = 4.16IVRR117 pKa = 11.84GGFSAGTFSLQALYY131 pKa = 9.61MEE133 pKa = 5.38HH134 pKa = 6.84LMCRR138 pKa = 11.84NRR140 pKa = 11.84WSQSNCGFDD149 pKa = 3.28LASYY153 pKa = 10.31RR154 pKa = 11.84GTKK157 pKa = 10.09LYY159 pKa = 9.93FVPHH163 pKa = 6.85PNYY166 pKa = 10.77DD167 pKa = 4.12YY168 pKa = 11.1IVLIDD173 pKa = 3.4PEE175 pKa = 4.31YY176 pKa = 11.01RR177 pKa = 11.84KK178 pKa = 10.14FEE180 pKa = 3.91QWIKK184 pKa = 10.76QPMHH188 pKa = 6.55PGVLITHH195 pKa = 6.17PQSRR199 pKa = 11.84IIRR202 pKa = 11.84SITHH206 pKa = 6.37AGPRR210 pKa = 11.84RR211 pKa = 11.84KK212 pKa = 8.68MPKK215 pKa = 8.94MFVPPPSTMNSGWQWMGEE233 pKa = 4.03LANEE237 pKa = 4.3GLFAFYY243 pKa = 10.2VQWIDD248 pKa = 5.72LGAPWLGDD256 pKa = 3.3VQNPNQVKK264 pKa = 8.2WWKK267 pKa = 9.23TGDD270 pKa = 3.42ATTKK274 pKa = 10.19PDD276 pKa = 3.25WVTKK280 pKa = 9.9GIEE283 pKa = 4.16MQGKK287 pKa = 6.7TVQEE291 pKa = 4.21GLDD294 pKa = 3.26AYY296 pKa = 8.81YY297 pKa = 9.31TQAGQGGKK305 pKa = 9.51KK306 pKa = 8.22WVNPGGAPYY315 pKa = 10.5QLGFGPFVFKK325 pKa = 10.71GSHH328 pKa = 6.15SGDD331 pKa = 3.41QEE333 pKa = 4.54HH334 pKa = 6.55YY335 pKa = 9.54PQICFFYY342 pKa = 10.47KK343 pKa = 10.46SYY345 pKa = 8.74WQWGGSTTSIKK356 pKa = 10.46NVCDD360 pKa = 3.8PKK362 pKa = 11.2DD363 pKa = 3.79NPNAEE368 pKa = 4.44YY369 pKa = 10.78LSAWNRR375 pKa = 11.84AGFQPAQARR384 pKa = 11.84NNDD387 pKa = 3.88IYY389 pKa = 11.22NQPP392 pKa = 3.25
Molecular weight: 46.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
749 |
89 |
392 |
249.7 |
28.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.142 ± 1.274 | 1.602 ± 0.376 |
4.406 ± 0.937 | 4.94 ± 1.557 |
3.605 ± 0.897 | 8.411 ± 1.33 |
2.27 ± 0.38 | 3.738 ± 0.31 |
4.806 ± 0.522 | 6.409 ± 1.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.537 ± 0.189 | 3.338 ± 0.598 |
6.943 ± 0.73 | 5.607 ± 0.378 |
9.613 ± 1.813 | 7.61 ± 2.165 |
6.943 ± 1.454 | 3.872 ± 0.4 |
3.204 ± 1.413 | 4.005 ± 1.524 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
