
Potato leafroll virus (strain Potato/Scotland/strain 1/1984) (PLrV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; Potato leafroll virus
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
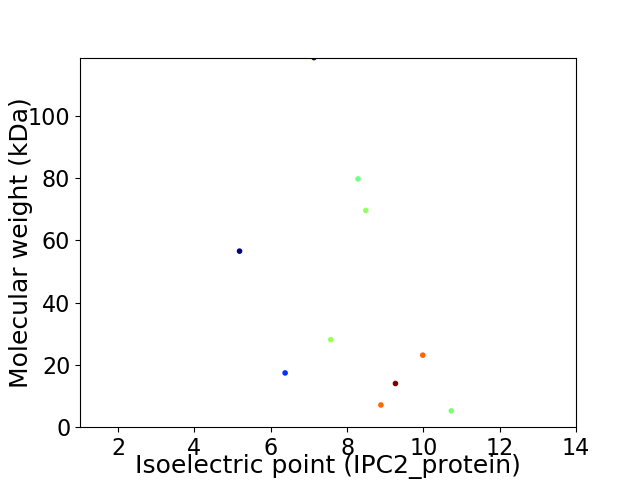
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
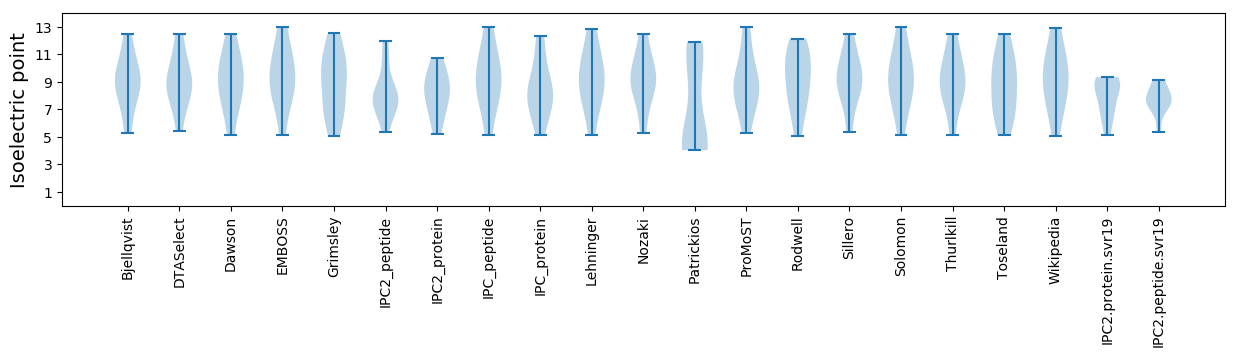
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P17525-2|MCAPS-2_PLRV1 Isoform of P17525 Isoform 2 of Minor capsid protein P3-RTD OS=Potato leafroll virus (strain Potato/Scotland/strain 1/1984) OX=12046 GN=ORF3/ORF5 PE=4 SV=2
VV1 pKa = 7.71DD2 pKa = 4.22SGSEE6 pKa = 4.06PSPSPQPTPTPTPQKK21 pKa = 9.54HH22 pKa = 5.31EE23 pKa = 4.05RR24 pKa = 11.84FIAYY28 pKa = 9.71VGIPMLTIQARR39 pKa = 11.84EE40 pKa = 3.85NDD42 pKa = 3.67DD43 pKa = 3.94QIILGSLGSQRR54 pKa = 11.84MKK56 pKa = 10.93YY57 pKa = 9.9IEE59 pKa = 5.51DD60 pKa = 3.36EE61 pKa = 4.18NQNYY65 pKa = 8.21TKK67 pKa = 10.51FSSEE71 pKa = 4.59YY72 pKa = 8.59YY73 pKa = 9.92SQSSMQAVPMYY84 pKa = 9.89YY85 pKa = 10.3FNVPKK90 pKa = 10.36GQWSVDD96 pKa = 3.39ISCEE100 pKa = 4.18GYY102 pKa = 10.21QPTSSTSDD110 pKa = 3.55PNRR113 pKa = 11.84GRR115 pKa = 11.84SDD117 pKa = 3.4GMIAYY122 pKa = 10.35SNADD126 pKa = 2.79SDD128 pKa = 4.02YY129 pKa = 9.5WNVGEE134 pKa = 4.53ADD136 pKa = 3.49GVKK139 pKa = 9.88ISKK142 pKa = 9.59LRR144 pKa = 11.84NDD146 pKa = 2.86NTYY149 pKa = 10.53RR150 pKa = 11.84QGHH153 pKa = 6.79PEE155 pKa = 4.05LEE157 pKa = 4.1INSCHH162 pKa = 5.84FRR164 pKa = 11.84EE165 pKa = 4.68GQLLEE170 pKa = 4.26RR171 pKa = 11.84DD172 pKa = 3.63ATISFHH178 pKa = 8.31VEE180 pKa = 3.32APTDD184 pKa = 3.4GRR186 pKa = 11.84FFLVGPAIQKK196 pKa = 5.49TAKK199 pKa = 9.33YY200 pKa = 9.73NYY202 pKa = 8.05TISYY206 pKa = 10.17GDD208 pKa = 3.06WTDD211 pKa = 3.7RR212 pKa = 11.84DD213 pKa = 3.91MEE215 pKa = 4.99LGLITVVLDD224 pKa = 3.29EE225 pKa = 4.77HH226 pKa = 8.61LEE228 pKa = 4.34GTGSANRR235 pKa = 11.84VRR237 pKa = 11.84RR238 pKa = 11.84PPRR241 pKa = 11.84EE242 pKa = 3.32GHH244 pKa = 6.13TYY246 pKa = 8.54MASPHH251 pKa = 5.77EE252 pKa = 4.38PEE254 pKa = 4.9GKK256 pKa = 9.47PVGNKK261 pKa = 9.3PRR263 pKa = 11.84DD264 pKa = 3.45EE265 pKa = 4.47TPIQTQEE272 pKa = 4.03RR273 pKa = 11.84QPDD276 pKa = 3.65QTPSDD281 pKa = 4.17DD282 pKa = 3.7VSDD285 pKa = 4.08AGSVNSGGPTEE296 pKa = 4.13SLRR299 pKa = 11.84LEE301 pKa = 4.24FGVNSDD307 pKa = 3.13STYY310 pKa = 11.27DD311 pKa = 3.5ATVDD315 pKa = 3.44GTDD318 pKa = 3.38WPRR321 pKa = 11.84IPPPRR326 pKa = 11.84HH327 pKa = 5.24PPEE330 pKa = 4.29PRR332 pKa = 11.84VSGNSRR338 pKa = 11.84TVTDD342 pKa = 5.0FSSKK346 pKa = 11.18ADD348 pKa = 4.0LLEE351 pKa = 4.27NWDD354 pKa = 4.58AEE356 pKa = 4.39HH357 pKa = 7.23FDD359 pKa = 3.6PGYY362 pKa = 10.6SKK364 pKa = 11.2EE365 pKa = 4.24DD366 pKa = 3.24VAAATIIAHH375 pKa = 6.99GSIQDD380 pKa = 3.53GRR382 pKa = 11.84SMLEE386 pKa = 3.66KK387 pKa = 10.35RR388 pKa = 11.84EE389 pKa = 4.03EE390 pKa = 3.98NVKK393 pKa = 10.85NKK395 pKa = 8.99TSSWKK400 pKa = 9.93PPSLKK405 pKa = 10.48AVSPAIAKK413 pKa = 9.51LRR415 pKa = 11.84SIRR418 pKa = 11.84KK419 pKa = 7.69SQPLEE424 pKa = 4.04GGTLNKK430 pKa = 10.38DD431 pKa = 3.0ATDD434 pKa = 4.12GVSSIGSGSLTGGTLKK450 pKa = 10.59RR451 pKa = 11.84KK452 pKa = 8.53ATIEE456 pKa = 3.84EE457 pKa = 4.38RR458 pKa = 11.84LLQTLTTEE466 pKa = 3.67QRR468 pKa = 11.84LWYY471 pKa = 10.26EE472 pKa = 4.08NFKK475 pKa = 9.32KK476 pKa = 9.31TNPPAATQWLFEE488 pKa = 4.28YY489 pKa = 10.49QPPPQVDD496 pKa = 3.24RR497 pKa = 11.84NIAEE501 pKa = 4.33KK502 pKa = 10.39PFQGRR507 pKa = 11.84KK508 pKa = 8.39
VV1 pKa = 7.71DD2 pKa = 4.22SGSEE6 pKa = 4.06PSPSPQPTPTPTPQKK21 pKa = 9.54HH22 pKa = 5.31EE23 pKa = 4.05RR24 pKa = 11.84FIAYY28 pKa = 9.71VGIPMLTIQARR39 pKa = 11.84EE40 pKa = 3.85NDD42 pKa = 3.67DD43 pKa = 3.94QIILGSLGSQRR54 pKa = 11.84MKK56 pKa = 10.93YY57 pKa = 9.9IEE59 pKa = 5.51DD60 pKa = 3.36EE61 pKa = 4.18NQNYY65 pKa = 8.21TKK67 pKa = 10.51FSSEE71 pKa = 4.59YY72 pKa = 8.59YY73 pKa = 9.92SQSSMQAVPMYY84 pKa = 9.89YY85 pKa = 10.3FNVPKK90 pKa = 10.36GQWSVDD96 pKa = 3.39ISCEE100 pKa = 4.18GYY102 pKa = 10.21QPTSSTSDD110 pKa = 3.55PNRR113 pKa = 11.84GRR115 pKa = 11.84SDD117 pKa = 3.4GMIAYY122 pKa = 10.35SNADD126 pKa = 2.79SDD128 pKa = 4.02YY129 pKa = 9.5WNVGEE134 pKa = 4.53ADD136 pKa = 3.49GVKK139 pKa = 9.88ISKK142 pKa = 9.59LRR144 pKa = 11.84NDD146 pKa = 2.86NTYY149 pKa = 10.53RR150 pKa = 11.84QGHH153 pKa = 6.79PEE155 pKa = 4.05LEE157 pKa = 4.1INSCHH162 pKa = 5.84FRR164 pKa = 11.84EE165 pKa = 4.68GQLLEE170 pKa = 4.26RR171 pKa = 11.84DD172 pKa = 3.63ATISFHH178 pKa = 8.31VEE180 pKa = 3.32APTDD184 pKa = 3.4GRR186 pKa = 11.84FFLVGPAIQKK196 pKa = 5.49TAKK199 pKa = 9.33YY200 pKa = 9.73NYY202 pKa = 8.05TISYY206 pKa = 10.17GDD208 pKa = 3.06WTDD211 pKa = 3.7RR212 pKa = 11.84DD213 pKa = 3.91MEE215 pKa = 4.99LGLITVVLDD224 pKa = 3.29EE225 pKa = 4.77HH226 pKa = 8.61LEE228 pKa = 4.34GTGSANRR235 pKa = 11.84VRR237 pKa = 11.84RR238 pKa = 11.84PPRR241 pKa = 11.84EE242 pKa = 3.32GHH244 pKa = 6.13TYY246 pKa = 8.54MASPHH251 pKa = 5.77EE252 pKa = 4.38PEE254 pKa = 4.9GKK256 pKa = 9.47PVGNKK261 pKa = 9.3PRR263 pKa = 11.84DD264 pKa = 3.45EE265 pKa = 4.47TPIQTQEE272 pKa = 4.03RR273 pKa = 11.84QPDD276 pKa = 3.65QTPSDD281 pKa = 4.17DD282 pKa = 3.7VSDD285 pKa = 4.08AGSVNSGGPTEE296 pKa = 4.13SLRR299 pKa = 11.84LEE301 pKa = 4.24FGVNSDD307 pKa = 3.13STYY310 pKa = 11.27DD311 pKa = 3.5ATVDD315 pKa = 3.44GTDD318 pKa = 3.38WPRR321 pKa = 11.84IPPPRR326 pKa = 11.84HH327 pKa = 5.24PPEE330 pKa = 4.29PRR332 pKa = 11.84VSGNSRR338 pKa = 11.84TVTDD342 pKa = 5.0FSSKK346 pKa = 11.18ADD348 pKa = 4.0LLEE351 pKa = 4.27NWDD354 pKa = 4.58AEE356 pKa = 4.39HH357 pKa = 7.23FDD359 pKa = 3.6PGYY362 pKa = 10.6SKK364 pKa = 11.2EE365 pKa = 4.24DD366 pKa = 3.24VAAATIIAHH375 pKa = 6.99GSIQDD380 pKa = 3.53GRR382 pKa = 11.84SMLEE386 pKa = 3.66KK387 pKa = 10.35RR388 pKa = 11.84EE389 pKa = 4.03EE390 pKa = 3.98NVKK393 pKa = 10.85NKK395 pKa = 8.99TSSWKK400 pKa = 9.93PPSLKK405 pKa = 10.48AVSPAIAKK413 pKa = 9.51LRR415 pKa = 11.84SIRR418 pKa = 11.84KK419 pKa = 7.69SQPLEE424 pKa = 4.04GGTLNKK430 pKa = 10.38DD431 pKa = 3.0ATDD434 pKa = 4.12GVSSIGSGSLTGGTLKK450 pKa = 10.59RR451 pKa = 11.84KK452 pKa = 8.53ATIEE456 pKa = 3.84EE457 pKa = 4.38RR458 pKa = 11.84LLQTLTTEE466 pKa = 3.67QRR468 pKa = 11.84LWYY471 pKa = 10.26EE472 pKa = 4.08NFKK475 pKa = 9.32KK476 pKa = 9.31TNPPAATQWLFEE488 pKa = 4.28YY489 pKa = 10.49QPPPQVDD496 pKa = 3.24RR497 pKa = 11.84NIAEE501 pKa = 4.33KK502 pKa = 10.39PFQGRR507 pKa = 11.84KK508 pKa = 8.39
Molecular weight: 56.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C769|P6_PLRV1 Uncharacterized protein P6 OS=Potato leafroll virus (strain Potato/Scotland/strain 1/1984) OX=12046 PE=4 SV=1
MM1 pKa = 7.19TPMRR5 pKa = 11.84ITVWRR10 pKa = 11.84EE11 pKa = 3.06RR12 pKa = 11.84LQQMRR17 pKa = 11.84PQRR20 pKa = 11.84KK21 pKa = 8.48LLKK24 pKa = 8.83QTQQRR29 pKa = 11.84RR30 pKa = 11.84LLHH33 pKa = 5.63QLQQRR38 pKa = 11.84KK39 pKa = 8.55LLL41 pKa = 4.03
MM1 pKa = 7.19TPMRR5 pKa = 11.84ITVWRR10 pKa = 11.84EE11 pKa = 3.06RR12 pKa = 11.84LQQMRR17 pKa = 11.84PQRR20 pKa = 11.84KK21 pKa = 8.48LLKK24 pKa = 8.83QTQQRR29 pKa = 11.84RR30 pKa = 11.84LLHH33 pKa = 5.63QLQQRR38 pKa = 11.84KK39 pKa = 8.55LLL41 pKa = 4.03
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3764 |
41 |
1062 |
376.4 |
41.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.333 ± 0.892 | 1.328 ± 0.3 |
4.145 ± 0.554 | 6.376 ± 0.463 |
3.666 ± 0.332 | 6.961 ± 0.341 |
2.046 ± 0.203 | 4.171 ± 0.24 |
5.898 ± 0.489 | 8.502 ± 1.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.171 | 4.304 ± 0.179 |
6.35 ± 0.622 | 4.623 ± 0.537 |
6.27 ± 0.694 | 9.458 ± 0.49 |
6.536 ± 0.309 | 5.074 ± 0.341 |
1.86 ± 0.225 | 3.162 ± 0.169 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
