
Cellulophaga phage phi39:1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
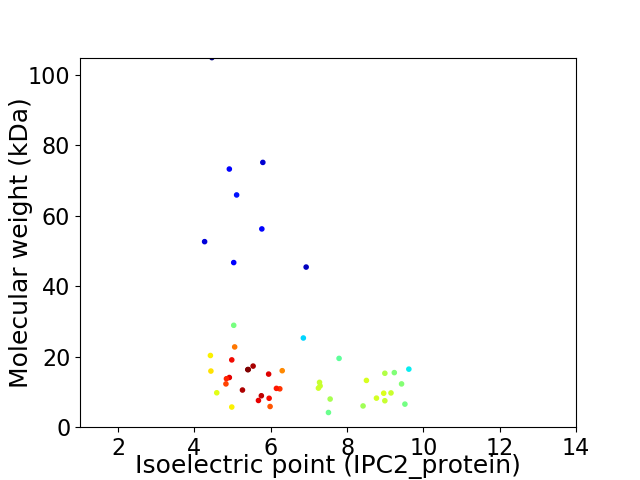
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0A2W2|S0A2W2_9CAUD DUF5675 domain-containing protein OS=Cellulophaga phage phi39:1 OX=1327993 GN=Phi39:1_gp30 PE=4 SV=1
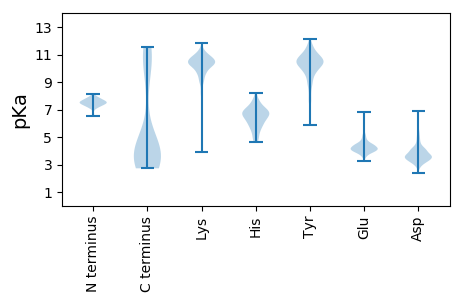
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 10.09LILFFGLIFCLEE15 pKa = 3.82MAAQVEE21 pKa = 4.39QVEE24 pKa = 4.45AVVLAKK30 pKa = 10.57KK31 pKa = 8.94STAEE35 pKa = 3.94LNALPSRR42 pKa = 11.84LKK44 pKa = 10.72QIGGIYY50 pKa = 10.14FDD52 pKa = 3.55VTLDD56 pKa = 6.03DD57 pKa = 4.05FVTWNGSNFVDD68 pKa = 5.46FSGEE72 pKa = 4.18TYY74 pKa = 10.46IFTGGISEE82 pKa = 4.49SGGTVTLEE90 pKa = 3.98NEE92 pKa = 3.7YY93 pKa = 9.11TAAEE97 pKa = 4.09KK98 pKa = 10.99SKK100 pKa = 11.04VSNLPNDD107 pKa = 3.34QSAVNEE113 pKa = 4.36TTVKK117 pKa = 10.33FDD119 pKa = 3.82KK120 pKa = 11.34NNANTFKK127 pKa = 10.7IKK129 pKa = 9.62TADD132 pKa = 3.54DD133 pKa = 3.76DD134 pKa = 4.06AAADD138 pKa = 4.01VILTINDD145 pKa = 3.68DD146 pKa = 3.77DD147 pKa = 4.28TPGGVEE153 pKa = 4.0YY154 pKa = 10.45FDD156 pKa = 4.78EE157 pKa = 4.52YY158 pKa = 11.55VEE160 pKa = 4.67EE161 pKa = 4.08ISGAVSDD168 pKa = 4.53GNLTITLGRR177 pKa = 11.84TNLSDD182 pKa = 5.27LISAAIPLPSPTGSGSYY199 pKa = 10.09SAGTGLSLGGSVFSLAPSVQSDD221 pKa = 3.21IDD223 pKa = 3.84SKK225 pKa = 11.4LNKK228 pKa = 10.2GLTNLGGTLLDD239 pKa = 3.73VKK241 pKa = 10.82SDD243 pKa = 3.21VGFRR247 pKa = 11.84FQPNEE252 pKa = 3.94GLSGFLVTPTLTSFLMGTADD272 pKa = 3.9LSKK275 pKa = 8.39MTSFSLTADD284 pKa = 3.41YY285 pKa = 10.85FRR287 pKa = 11.84TNLSDD292 pKa = 4.55AKK294 pKa = 10.54ILAQGDD300 pKa = 4.07DD301 pKa = 3.27SMLTYY306 pKa = 10.95GFVKK310 pKa = 10.56NYY312 pKa = 10.28FLTKK316 pKa = 10.17DD317 pKa = 3.21AAAALYY323 pKa = 10.5APIGSSTGGGDD334 pKa = 4.37VDD336 pKa = 5.94DD337 pKa = 6.09ADD339 pKa = 4.87ADD341 pKa = 4.12PLNEE345 pKa = 3.9IQTLSVSGNTVTLSKK360 pKa = 11.06DD361 pKa = 2.61GGAFVIPSGGINQTIGNSVLTLKK384 pKa = 11.05GITQGDD390 pKa = 3.51ITTTAKK396 pKa = 9.91WSLTGSVLTLHH407 pKa = 6.26YY408 pKa = 10.15RR409 pKa = 11.84FVMSGNKK416 pKa = 6.25TTNGLVTIDD425 pKa = 3.57GLPYY429 pKa = 9.76RR430 pKa = 11.84PEE432 pKa = 4.34GLTHH436 pKa = 6.63HH437 pKa = 6.9AMFITGSTSNSMIGAFTDD455 pKa = 3.57ANYY458 pKa = 11.17SNGNVLRR465 pKa = 11.84PFVYY469 pKa = 10.55LSGGNNPIQLNTGQSLNSGTQILGTVTYY497 pKa = 10.22IINN500 pKa = 3.55
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 10.09LILFFGLIFCLEE15 pKa = 3.82MAAQVEE21 pKa = 4.39QVEE24 pKa = 4.45AVVLAKK30 pKa = 10.57KK31 pKa = 8.94STAEE35 pKa = 3.94LNALPSRR42 pKa = 11.84LKK44 pKa = 10.72QIGGIYY50 pKa = 10.14FDD52 pKa = 3.55VTLDD56 pKa = 6.03DD57 pKa = 4.05FVTWNGSNFVDD68 pKa = 5.46FSGEE72 pKa = 4.18TYY74 pKa = 10.46IFTGGISEE82 pKa = 4.49SGGTVTLEE90 pKa = 3.98NEE92 pKa = 3.7YY93 pKa = 9.11TAAEE97 pKa = 4.09KK98 pKa = 10.99SKK100 pKa = 11.04VSNLPNDD107 pKa = 3.34QSAVNEE113 pKa = 4.36TTVKK117 pKa = 10.33FDD119 pKa = 3.82KK120 pKa = 11.34NNANTFKK127 pKa = 10.7IKK129 pKa = 9.62TADD132 pKa = 3.54DD133 pKa = 3.76DD134 pKa = 4.06AAADD138 pKa = 4.01VILTINDD145 pKa = 3.68DD146 pKa = 3.77DD147 pKa = 4.28TPGGVEE153 pKa = 4.0YY154 pKa = 10.45FDD156 pKa = 4.78EE157 pKa = 4.52YY158 pKa = 11.55VEE160 pKa = 4.67EE161 pKa = 4.08ISGAVSDD168 pKa = 4.53GNLTITLGRR177 pKa = 11.84TNLSDD182 pKa = 5.27LISAAIPLPSPTGSGSYY199 pKa = 10.09SAGTGLSLGGSVFSLAPSVQSDD221 pKa = 3.21IDD223 pKa = 3.84SKK225 pKa = 11.4LNKK228 pKa = 10.2GLTNLGGTLLDD239 pKa = 3.73VKK241 pKa = 10.82SDD243 pKa = 3.21VGFRR247 pKa = 11.84FQPNEE252 pKa = 3.94GLSGFLVTPTLTSFLMGTADD272 pKa = 3.9LSKK275 pKa = 8.39MTSFSLTADD284 pKa = 3.41YY285 pKa = 10.85FRR287 pKa = 11.84TNLSDD292 pKa = 4.55AKK294 pKa = 10.54ILAQGDD300 pKa = 4.07DD301 pKa = 3.27SMLTYY306 pKa = 10.95GFVKK310 pKa = 10.56NYY312 pKa = 10.28FLTKK316 pKa = 10.17DD317 pKa = 3.21AAAALYY323 pKa = 10.5APIGSSTGGGDD334 pKa = 4.37VDD336 pKa = 5.94DD337 pKa = 6.09ADD339 pKa = 4.87ADD341 pKa = 4.12PLNEE345 pKa = 3.9IQTLSVSGNTVTLSKK360 pKa = 11.06DD361 pKa = 2.61GGAFVIPSGGINQTIGNSVLTLKK384 pKa = 11.05GITQGDD390 pKa = 3.51ITTTAKK396 pKa = 9.91WSLTGSVLTLHH407 pKa = 6.26YY408 pKa = 10.15RR409 pKa = 11.84FVMSGNKK416 pKa = 6.25TTNGLVTIDD425 pKa = 3.57GLPYY429 pKa = 9.76RR430 pKa = 11.84PEE432 pKa = 4.34GLTHH436 pKa = 6.63HH437 pKa = 6.9AMFITGSTSNSMIGAFTDD455 pKa = 3.57ANYY458 pKa = 11.17SNGNVLRR465 pKa = 11.84PFVYY469 pKa = 10.55LSGGNNPIQLNTGQSLNSGTQILGTVTYY497 pKa = 10.22IINN500 pKa = 3.55
Molecular weight: 52.68 kDa
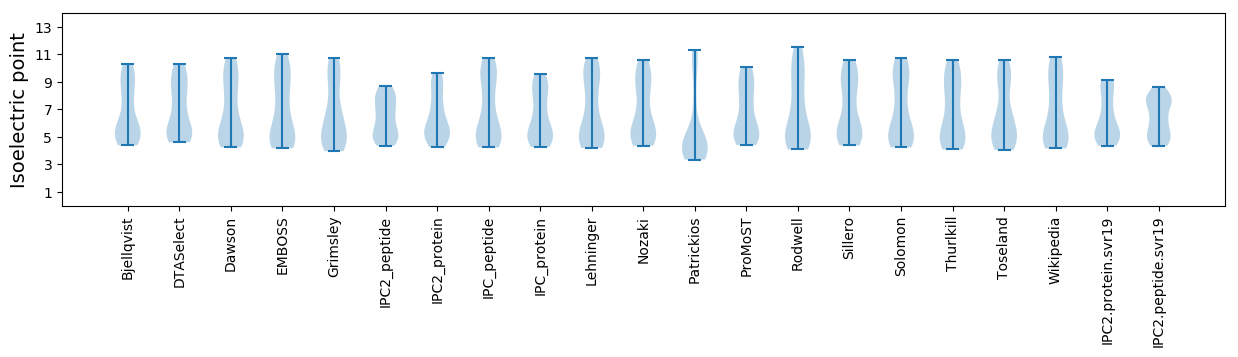
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0A2X0|S0A2X0_9CAUD Uncharacterized protein OS=Cellulophaga phage phi39:1 OX=1327993 GN=Phi39:1_gp45 PE=4 SV=1
MM1 pKa = 7.31TKK3 pKa = 10.6NIILVIVLFSVVSSYY18 pKa = 11.6NLSKK22 pKa = 11.17YY23 pKa = 10.64SDD25 pKa = 5.16GILHH29 pKa = 6.73WKK31 pKa = 10.25SFIPLFFYY39 pKa = 10.99CFLLSVFVVVFFKK52 pKa = 10.89ILVKK56 pKa = 10.41LCKK59 pKa = 10.29SILNGII65 pKa = 4.24
MM1 pKa = 7.31TKK3 pKa = 10.6NIILVIVLFSVVSSYY18 pKa = 11.6NLSKK22 pKa = 11.17YY23 pKa = 10.64SDD25 pKa = 5.16GILHH29 pKa = 6.73WKK31 pKa = 10.25SFIPLFFYY39 pKa = 10.99CFLLSVFVVVFFKK52 pKa = 10.89ILVKK56 pKa = 10.41LCKK59 pKa = 10.29SILNGII65 pKa = 4.24
Molecular weight: 7.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9341 |
33 |
935 |
194.6 |
21.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.905 ± 0.708 | 0.899 ± 0.18 |
6.306 ± 0.318 | 6.263 ± 0.417 |
4.989 ± 0.278 | 7.034 ± 0.561 |
1.092 ± 0.166 | 7.601 ± 0.317 |
8.222 ± 0.537 | 9.175 ± 0.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.916 ± 0.186 | 6.188 ± 0.355 |
2.826 ± 0.177 | 2.858 ± 0.204 |
3.287 ± 0.207 | 7.365 ± 0.417 |
5.417 ± 0.461 | 7.034 ± 0.379 |
1.028 ± 0.113 | 3.597 ± 0.396 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
