
Wenling tombus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.77
Get precalculated fractions of proteins
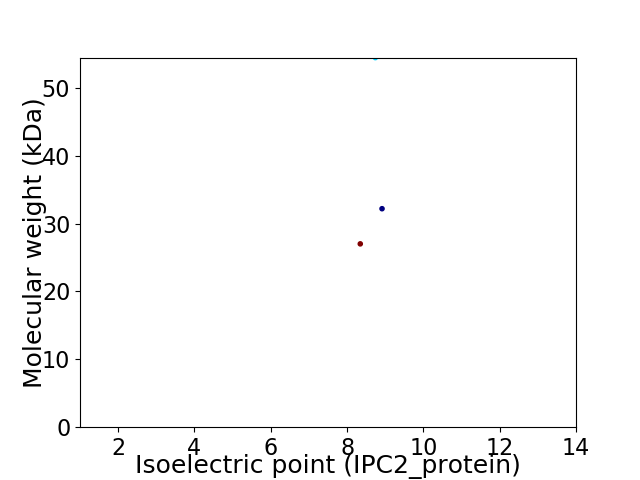
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
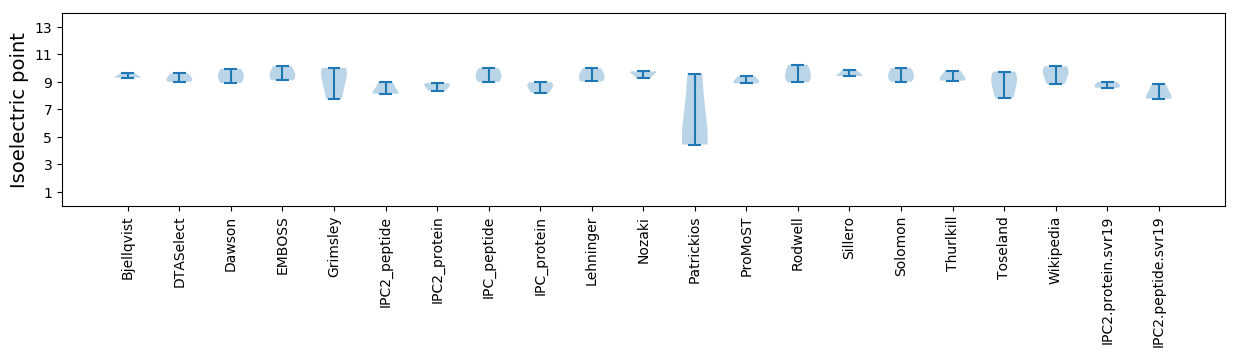
Protein with the lowest isoelectric point:
>tr|A0A1L3KGT1|A0A1L3KGT1_9VIRU Uncharacterized protein OS=Wenling tombus-like virus 3 OX=1923545 PE=4 SV=1
MM1 pKa = 7.38VILAEE6 pKa = 4.84DD7 pKa = 3.92EE8 pKa = 4.5TKK10 pKa = 10.69SGCGHH15 pKa = 6.03SCRR18 pKa = 11.84TPRR21 pKa = 11.84CTRR24 pKa = 11.84TRR26 pKa = 11.84SGPINGSASCGRR38 pKa = 11.84DD39 pKa = 3.11TSGSRR44 pKa = 11.84HH45 pKa = 5.19SNRR48 pKa = 11.84HH49 pKa = 4.05RR50 pKa = 11.84FRR52 pKa = 11.84YY53 pKa = 8.7RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84GKK59 pKa = 9.24PRR61 pKa = 11.84CPRR64 pKa = 11.84CRR66 pKa = 11.84HH67 pKa = 5.99RR68 pKa = 11.84GDD70 pKa = 3.22GKK72 pKa = 10.75KK73 pKa = 9.89FCVSQRR79 pKa = 11.84CASISATEE87 pKa = 4.14KK88 pKa = 10.58DD89 pKa = 3.56QSILSEE95 pKa = 4.83DD96 pKa = 3.79GMDD99 pKa = 3.25AHH101 pKa = 7.39AGTSCTHH108 pKa = 6.45SGPAKK113 pKa = 9.81TYY115 pKa = 11.1AEE117 pKa = 4.65LSTHH121 pKa = 6.23SSWALPGDD129 pKa = 4.13CGNVAKK135 pKa = 10.53EE136 pKa = 4.04LVQSNSRR143 pKa = 11.84CNIIRR148 pKa = 11.84TPGCGSTTDD157 pKa = 4.22RR158 pKa = 11.84RR159 pKa = 11.84DD160 pKa = 3.45HH161 pKa = 6.84FCGQSGGSDD170 pKa = 3.54VVLKK174 pKa = 10.45AASSVSPSDD183 pKa = 3.42KK184 pKa = 10.49TPLIHH189 pKa = 7.43VSGKK193 pKa = 9.32SEE195 pKa = 3.89EE196 pKa = 4.03VLGRR200 pKa = 11.84FRR202 pKa = 11.84LAASHH207 pKa = 7.07AARR210 pKa = 11.84NLFRR214 pKa = 11.84HH215 pKa = 5.85LTLCCGGIYY224 pKa = 10.28ACRR227 pKa = 11.84EE228 pKa = 4.07RR229 pKa = 11.84VGGAQLDD236 pKa = 4.17DD237 pKa = 3.88SCEE240 pKa = 3.82EE241 pKa = 4.29GLQSVHH247 pKa = 6.37QSRR250 pKa = 4.07
MM1 pKa = 7.38VILAEE6 pKa = 4.84DD7 pKa = 3.92EE8 pKa = 4.5TKK10 pKa = 10.69SGCGHH15 pKa = 6.03SCRR18 pKa = 11.84TPRR21 pKa = 11.84CTRR24 pKa = 11.84TRR26 pKa = 11.84SGPINGSASCGRR38 pKa = 11.84DD39 pKa = 3.11TSGSRR44 pKa = 11.84HH45 pKa = 5.19SNRR48 pKa = 11.84HH49 pKa = 4.05RR50 pKa = 11.84FRR52 pKa = 11.84YY53 pKa = 8.7RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84GKK59 pKa = 9.24PRR61 pKa = 11.84CPRR64 pKa = 11.84CRR66 pKa = 11.84HH67 pKa = 5.99RR68 pKa = 11.84GDD70 pKa = 3.22GKK72 pKa = 10.75KK73 pKa = 9.89FCVSQRR79 pKa = 11.84CASISATEE87 pKa = 4.14KK88 pKa = 10.58DD89 pKa = 3.56QSILSEE95 pKa = 4.83DD96 pKa = 3.79GMDD99 pKa = 3.25AHH101 pKa = 7.39AGTSCTHH108 pKa = 6.45SGPAKK113 pKa = 9.81TYY115 pKa = 11.1AEE117 pKa = 4.65LSTHH121 pKa = 6.23SSWALPGDD129 pKa = 4.13CGNVAKK135 pKa = 10.53EE136 pKa = 4.04LVQSNSRR143 pKa = 11.84CNIIRR148 pKa = 11.84TPGCGSTTDD157 pKa = 4.22RR158 pKa = 11.84RR159 pKa = 11.84DD160 pKa = 3.45HH161 pKa = 6.84FCGQSGGSDD170 pKa = 3.54VVLKK174 pKa = 10.45AASSVSPSDD183 pKa = 3.42KK184 pKa = 10.49TPLIHH189 pKa = 7.43VSGKK193 pKa = 9.32SEE195 pKa = 3.89EE196 pKa = 4.03VLGRR200 pKa = 11.84FRR202 pKa = 11.84LAASHH207 pKa = 7.07AARR210 pKa = 11.84NLFRR214 pKa = 11.84HH215 pKa = 5.85LTLCCGGIYY224 pKa = 10.28ACRR227 pKa = 11.84EE228 pKa = 4.07RR229 pKa = 11.84VGGAQLDD236 pKa = 4.17DD237 pKa = 3.88SCEE240 pKa = 3.82EE241 pKa = 4.29GLQSVHH247 pKa = 6.37QSRR250 pKa = 4.07
Molecular weight: 27.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH11|A0A1L3KH11_9VIRU RNA-directed RNA polymerase OS=Wenling tombus-like virus 3 OX=1923545 PE=4 SV=1
MM1 pKa = 6.91KK2 pKa = 8.97TWSGGICVMPSFHH15 pKa = 6.66TKK17 pKa = 9.2QVGPRR22 pKa = 11.84TNYY25 pKa = 9.51RR26 pKa = 11.84WSFWQRR32 pKa = 11.84MKK34 pKa = 10.89RR35 pKa = 11.84SLDD38 pKa = 2.78VDD40 pKa = 4.17TPVEE44 pKa = 3.87PHH46 pKa = 7.0DD47 pKa = 5.12ALGQGPAPLMGVRR60 pKa = 11.84PVDD63 pKa = 4.84EE64 pKa = 4.94IPVDD68 pKa = 3.63QDD70 pKa = 3.17IPIATASGTVGVEE83 pKa = 4.16VNPGAHH89 pKa = 7.06DD90 pKa = 3.7AAIAATGRR98 pKa = 11.84SFAFHH103 pKa = 6.71NDD105 pKa = 2.03AHH107 pKa = 6.45RR108 pKa = 11.84FLRR111 pKa = 11.84QRR113 pKa = 11.84RR114 pKa = 11.84INRR117 pKa = 11.84FSPKK121 pKa = 9.67TEE123 pKa = 3.55WTRR126 pKa = 11.84MRR128 pKa = 11.84VHH130 pKa = 7.18RR131 pKa = 11.84VPIQDD136 pKa = 3.35QRR138 pKa = 11.84KK139 pKa = 8.38PMPNYY144 pKa = 10.05RR145 pKa = 11.84PTAHH149 pKa = 7.33GLFQEE154 pKa = 4.48TAEE157 pKa = 4.09MLQKK161 pKa = 10.78NWFNPTLDD169 pKa = 3.43VTLSVPRR176 pKa = 11.84GVDD179 pKa = 2.82RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84IEE184 pKa = 4.56EE185 pKa = 4.03ITSVASPEE193 pKa = 3.93DD194 pKa = 3.58LTWFLRR200 pKa = 11.84RR201 pKa = 11.84QALFRR206 pKa = 11.84PRR208 pKa = 11.84TKK210 pKa = 10.64LLLSMLVEE218 pKa = 4.18KK219 pKa = 10.66AKK221 pKa = 10.88KK222 pKa = 10.13FLDD225 pKa = 4.26DD226 pKa = 3.27SDD228 pKa = 4.98LRR230 pKa = 11.84HH231 pKa = 5.19HH232 pKa = 6.15TQRR235 pKa = 11.84EE236 pKa = 4.36IYY238 pKa = 10.15FGTLHH243 pKa = 6.23SVAAAYY249 pKa = 8.82MPVEE253 pKa = 4.37RR254 pKa = 11.84EE255 pKa = 4.13LEE257 pKa = 4.13VLNWMTAAKK266 pKa = 9.91KK267 pKa = 10.38DD268 pKa = 3.78YY269 pKa = 10.61NQFISPDD276 pKa = 3.75DD277 pKa = 4.11ASSS280 pKa = 2.89
MM1 pKa = 6.91KK2 pKa = 8.97TWSGGICVMPSFHH15 pKa = 6.66TKK17 pKa = 9.2QVGPRR22 pKa = 11.84TNYY25 pKa = 9.51RR26 pKa = 11.84WSFWQRR32 pKa = 11.84MKK34 pKa = 10.89RR35 pKa = 11.84SLDD38 pKa = 2.78VDD40 pKa = 4.17TPVEE44 pKa = 3.87PHH46 pKa = 7.0DD47 pKa = 5.12ALGQGPAPLMGVRR60 pKa = 11.84PVDD63 pKa = 4.84EE64 pKa = 4.94IPVDD68 pKa = 3.63QDD70 pKa = 3.17IPIATASGTVGVEE83 pKa = 4.16VNPGAHH89 pKa = 7.06DD90 pKa = 3.7AAIAATGRR98 pKa = 11.84SFAFHH103 pKa = 6.71NDD105 pKa = 2.03AHH107 pKa = 6.45RR108 pKa = 11.84FLRR111 pKa = 11.84QRR113 pKa = 11.84RR114 pKa = 11.84INRR117 pKa = 11.84FSPKK121 pKa = 9.67TEE123 pKa = 3.55WTRR126 pKa = 11.84MRR128 pKa = 11.84VHH130 pKa = 7.18RR131 pKa = 11.84VPIQDD136 pKa = 3.35QRR138 pKa = 11.84KK139 pKa = 8.38PMPNYY144 pKa = 10.05RR145 pKa = 11.84PTAHH149 pKa = 7.33GLFQEE154 pKa = 4.48TAEE157 pKa = 4.09MLQKK161 pKa = 10.78NWFNPTLDD169 pKa = 3.43VTLSVPRR176 pKa = 11.84GVDD179 pKa = 2.82RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84IEE184 pKa = 4.56EE185 pKa = 4.03ITSVASPEE193 pKa = 3.93DD194 pKa = 3.58LTWFLRR200 pKa = 11.84RR201 pKa = 11.84QALFRR206 pKa = 11.84PRR208 pKa = 11.84TKK210 pKa = 10.64LLLSMLVEE218 pKa = 4.18KK219 pKa = 10.66AKK221 pKa = 10.88KK222 pKa = 10.13FLDD225 pKa = 4.26DD226 pKa = 3.27SDD228 pKa = 4.98LRR230 pKa = 11.84HH231 pKa = 5.19HH232 pKa = 6.15TQRR235 pKa = 11.84EE236 pKa = 4.36IYY238 pKa = 10.15FGTLHH243 pKa = 6.23SVAAAYY249 pKa = 8.82MPVEE253 pKa = 4.37RR254 pKa = 11.84EE255 pKa = 4.13LEE257 pKa = 4.13VLNWMTAAKK266 pKa = 9.91KK267 pKa = 10.38DD268 pKa = 3.78YY269 pKa = 10.61NQFISPDD276 pKa = 3.75DD277 pKa = 4.11ASSS280 pKa = 2.89
Molecular weight: 32.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
998 |
250 |
468 |
332.7 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.914 ± 0.443 | 2.505 ± 1.341 |
6.112 ± 0.287 | 4.609 ± 0.136 |
4.709 ± 0.992 | 6.513 ± 1.212 |
3.707 ± 0.37 | 4.208 ± 0.432 |
4.509 ± 0.242 | 8.317 ± 1.288 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.531 | 3.607 ± 0.534 |
5.21 ± 0.807 | 2.505 ± 0.779 |
9.82 ± 0.61 | 8.617 ± 1.467 |
4.509 ± 1.28 | 6.212 ± 0.567 |
1.603 ± 0.401 | 3.407 ± 1.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
