
Changjiang tombus-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.97
Get precalculated fractions of proteins
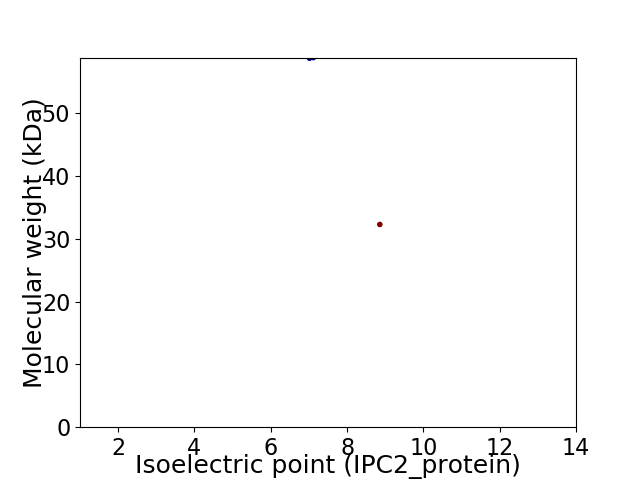
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFW7|A0A1L3KFW7_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 15 OX=1922808 PE=4 SV=1
MM1 pKa = 7.52PPGVEE6 pKa = 4.11CHH8 pKa = 6.92SLGLDD13 pKa = 2.83MKK15 pKa = 10.49GRR17 pKa = 11.84HH18 pKa = 5.05KK19 pKa = 10.61EE20 pKa = 3.7RR21 pKa = 11.84YY22 pKa = 9.3LVMDD26 pKa = 4.07TTRR29 pKa = 11.84PAAGLCFTHH38 pKa = 6.54NNSVPNILRR47 pKa = 11.84GLGEE51 pKa = 4.49RR52 pKa = 11.84LMMVPNDD59 pKa = 4.42DD60 pKa = 4.9GGFSPPPEE68 pKa = 3.88PTVFDD73 pKa = 3.82LGEE76 pKa = 3.85YY77 pKa = 10.2SKK79 pKa = 11.53AVLRR83 pKa = 11.84KK84 pKa = 8.45MPKK87 pKa = 8.94HH88 pKa = 6.06LEE90 pKa = 4.09PLEE93 pKa = 4.49HH94 pKa = 7.51DD95 pKa = 4.29EE96 pKa = 6.18FVMLYY101 pKa = 10.22DD102 pKa = 3.55GPKK105 pKa = 8.68RR106 pKa = 11.84KK107 pKa = 9.55RR108 pKa = 11.84YY109 pKa = 8.15EE110 pKa = 3.67AAARR114 pKa = 11.84ALLEE118 pKa = 4.36RR119 pKa = 11.84EE120 pKa = 4.46LDD122 pKa = 3.71AKK124 pKa = 10.65DD125 pKa = 3.29WQIKK129 pKa = 10.27LFIKK133 pKa = 10.7DD134 pKa = 4.13EE135 pKa = 4.5IVCSWAKK142 pKa = 10.65ADD144 pKa = 3.82PAPRR148 pKa = 11.84LISPRR153 pKa = 11.84SPEE156 pKa = 3.63YY157 pKa = 10.35CLEE160 pKa = 4.07VGCFIKK166 pKa = 10.59PIEE169 pKa = 4.33HH170 pKa = 7.15LLYY173 pKa = 10.69KK174 pKa = 10.52AVARR178 pKa = 11.84VWGEE182 pKa = 3.6TTIAKK187 pKa = 9.81GLNFNQRR194 pKa = 11.84GEE196 pKa = 4.23LIKK199 pKa = 10.68EE200 pKa = 3.8KK201 pKa = 10.06WNSFKK206 pKa = 11.16KK207 pKa = 9.99PVAVGLDD214 pKa = 3.18ASRR217 pKa = 11.84FDD219 pKa = 3.48QHH221 pKa = 7.45VSLSALQWEE230 pKa = 4.26HH231 pKa = 7.23SIYY234 pKa = 10.67LGCFPKK240 pKa = 10.24HH241 pKa = 5.85RR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84LQKK247 pKa = 10.69LLSKK251 pKa = 9.19QLRR254 pKa = 11.84NFGVCYY260 pKa = 10.58VDD262 pKa = 3.69DD263 pKa = 3.93HH264 pKa = 7.07RR265 pKa = 11.84ITYY268 pKa = 10.27SRR270 pKa = 11.84DD271 pKa = 2.97GGRR274 pKa = 11.84MSGDD278 pKa = 3.19MNTALGNCLIMTGLVWTYY296 pKa = 10.68AKK298 pKa = 10.26QRR300 pKa = 11.84GITVKK305 pKa = 10.73LINDD309 pKa = 3.58GDD311 pKa = 4.07DD312 pKa = 3.15CVVFMEE318 pKa = 4.96EE319 pKa = 3.74SDD321 pKa = 4.31LPAFMEE327 pKa = 5.24GLRR330 pKa = 11.84QWFLEE335 pKa = 3.79RR336 pKa = 11.84GFNMKK341 pKa = 9.7VEE343 pKa = 4.29QPAYY347 pKa = 8.68EE348 pKa = 4.05LEE350 pKa = 5.08AIEE353 pKa = 5.26FCQCHH358 pKa = 5.65PVFNGDD364 pKa = 3.2QYY366 pKa = 10.31TMCRR370 pKa = 11.84NIHH373 pKa = 6.18KK374 pKa = 10.72ALFTDD379 pKa = 4.21VAHH382 pKa = 6.75VGRR385 pKa = 11.84TAEE388 pKa = 4.27EE389 pKa = 3.57IRR391 pKa = 11.84GIRR394 pKa = 11.84EE395 pKa = 3.71AVALCGAAWSKK406 pKa = 10.65GLPIFPTFYY415 pKa = 10.2AGLRR419 pKa = 11.84TGHH422 pKa = 6.38KK423 pKa = 9.88PSVLRR428 pKa = 11.84HH429 pKa = 6.19SGTFWNSQGCKK440 pKa = 10.09SGTKK444 pKa = 9.97YY445 pKa = 8.46VTPRR449 pKa = 11.84ARR451 pKa = 11.84LSFEE455 pKa = 3.89RR456 pKa = 11.84AFGLSPSEE464 pKa = 3.71QVAIEE469 pKa = 4.22NFYY472 pKa = 11.17QSLPRR477 pKa = 11.84TSIDD481 pKa = 3.33EE482 pKa = 3.82PHH484 pKa = 6.6LVFEE488 pKa = 5.1HH489 pKa = 6.74SPTDD493 pKa = 3.44SSTHH497 pKa = 5.87HH498 pKa = 7.01PLFVSDD504 pKa = 3.89SLNYY508 pKa = 10.87LLFNHH513 pKa = 6.72GKK515 pKa = 9.99EE516 pKa = 4.23EE517 pKa = 4.0
MM1 pKa = 7.52PPGVEE6 pKa = 4.11CHH8 pKa = 6.92SLGLDD13 pKa = 2.83MKK15 pKa = 10.49GRR17 pKa = 11.84HH18 pKa = 5.05KK19 pKa = 10.61EE20 pKa = 3.7RR21 pKa = 11.84YY22 pKa = 9.3LVMDD26 pKa = 4.07TTRR29 pKa = 11.84PAAGLCFTHH38 pKa = 6.54NNSVPNILRR47 pKa = 11.84GLGEE51 pKa = 4.49RR52 pKa = 11.84LMMVPNDD59 pKa = 4.42DD60 pKa = 4.9GGFSPPPEE68 pKa = 3.88PTVFDD73 pKa = 3.82LGEE76 pKa = 3.85YY77 pKa = 10.2SKK79 pKa = 11.53AVLRR83 pKa = 11.84KK84 pKa = 8.45MPKK87 pKa = 8.94HH88 pKa = 6.06LEE90 pKa = 4.09PLEE93 pKa = 4.49HH94 pKa = 7.51DD95 pKa = 4.29EE96 pKa = 6.18FVMLYY101 pKa = 10.22DD102 pKa = 3.55GPKK105 pKa = 8.68RR106 pKa = 11.84KK107 pKa = 9.55RR108 pKa = 11.84YY109 pKa = 8.15EE110 pKa = 3.67AAARR114 pKa = 11.84ALLEE118 pKa = 4.36RR119 pKa = 11.84EE120 pKa = 4.46LDD122 pKa = 3.71AKK124 pKa = 10.65DD125 pKa = 3.29WQIKK129 pKa = 10.27LFIKK133 pKa = 10.7DD134 pKa = 4.13EE135 pKa = 4.5IVCSWAKK142 pKa = 10.65ADD144 pKa = 3.82PAPRR148 pKa = 11.84LISPRR153 pKa = 11.84SPEE156 pKa = 3.63YY157 pKa = 10.35CLEE160 pKa = 4.07VGCFIKK166 pKa = 10.59PIEE169 pKa = 4.33HH170 pKa = 7.15LLYY173 pKa = 10.69KK174 pKa = 10.52AVARR178 pKa = 11.84VWGEE182 pKa = 3.6TTIAKK187 pKa = 9.81GLNFNQRR194 pKa = 11.84GEE196 pKa = 4.23LIKK199 pKa = 10.68EE200 pKa = 3.8KK201 pKa = 10.06WNSFKK206 pKa = 11.16KK207 pKa = 9.99PVAVGLDD214 pKa = 3.18ASRR217 pKa = 11.84FDD219 pKa = 3.48QHH221 pKa = 7.45VSLSALQWEE230 pKa = 4.26HH231 pKa = 7.23SIYY234 pKa = 10.67LGCFPKK240 pKa = 10.24HH241 pKa = 5.85RR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84LQKK247 pKa = 10.69LLSKK251 pKa = 9.19QLRR254 pKa = 11.84NFGVCYY260 pKa = 10.58VDD262 pKa = 3.69DD263 pKa = 3.93HH264 pKa = 7.07RR265 pKa = 11.84ITYY268 pKa = 10.27SRR270 pKa = 11.84DD271 pKa = 2.97GGRR274 pKa = 11.84MSGDD278 pKa = 3.19MNTALGNCLIMTGLVWTYY296 pKa = 10.68AKK298 pKa = 10.26QRR300 pKa = 11.84GITVKK305 pKa = 10.73LINDD309 pKa = 3.58GDD311 pKa = 4.07DD312 pKa = 3.15CVVFMEE318 pKa = 4.96EE319 pKa = 3.74SDD321 pKa = 4.31LPAFMEE327 pKa = 5.24GLRR330 pKa = 11.84QWFLEE335 pKa = 3.79RR336 pKa = 11.84GFNMKK341 pKa = 9.7VEE343 pKa = 4.29QPAYY347 pKa = 8.68EE348 pKa = 4.05LEE350 pKa = 5.08AIEE353 pKa = 5.26FCQCHH358 pKa = 5.65PVFNGDD364 pKa = 3.2QYY366 pKa = 10.31TMCRR370 pKa = 11.84NIHH373 pKa = 6.18KK374 pKa = 10.72ALFTDD379 pKa = 4.21VAHH382 pKa = 6.75VGRR385 pKa = 11.84TAEE388 pKa = 4.27EE389 pKa = 3.57IRR391 pKa = 11.84GIRR394 pKa = 11.84EE395 pKa = 3.71AVALCGAAWSKK406 pKa = 10.65GLPIFPTFYY415 pKa = 10.2AGLRR419 pKa = 11.84TGHH422 pKa = 6.38KK423 pKa = 9.88PSVLRR428 pKa = 11.84HH429 pKa = 6.19SGTFWNSQGCKK440 pKa = 10.09SGTKK444 pKa = 9.97YY445 pKa = 8.46VTPRR449 pKa = 11.84ARR451 pKa = 11.84LSFEE455 pKa = 3.89RR456 pKa = 11.84AFGLSPSEE464 pKa = 3.71QVAIEE469 pKa = 4.22NFYY472 pKa = 11.17QSLPRR477 pKa = 11.84TSIDD481 pKa = 3.33EE482 pKa = 3.82PHH484 pKa = 6.6LVFEE488 pKa = 5.1HH489 pKa = 6.74SPTDD493 pKa = 3.44SSTHH497 pKa = 5.87HH498 pKa = 7.01PLFVSDD504 pKa = 3.89SLNYY508 pKa = 10.87LLFNHH513 pKa = 6.72GKK515 pKa = 9.99EE516 pKa = 4.23EE517 pKa = 4.0
Molecular weight: 58.8 kDa
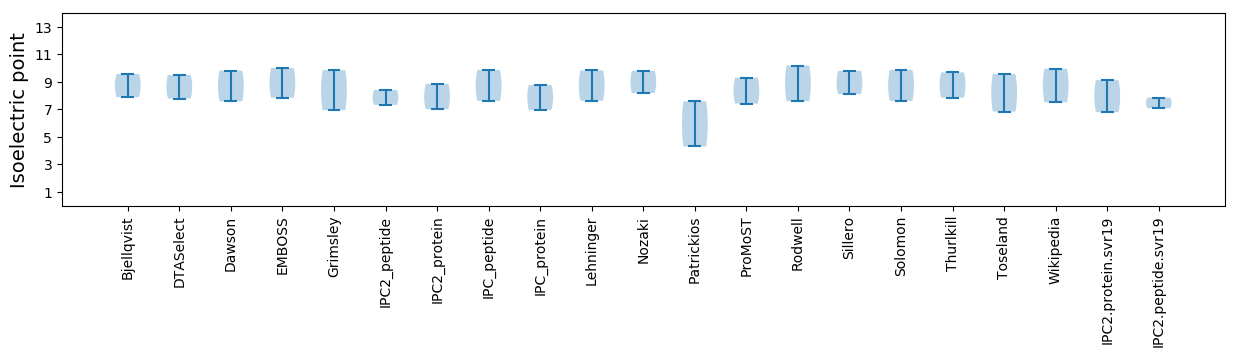
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFW7|A0A1L3KFW7_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 15 OX=1922808 PE=4 SV=1
MM1 pKa = 7.59AKK3 pKa = 9.98KK4 pKa = 10.29SNKK7 pKa = 9.39SPQVQKK13 pKa = 6.72TTRR16 pKa = 11.84PKK18 pKa = 9.0RR19 pKa = 11.84QRR21 pKa = 11.84KK22 pKa = 8.41GRR24 pKa = 11.84IAGVQGKK31 pKa = 9.12ALQLARR37 pKa = 11.84LLNDD41 pKa = 3.58PCSAEE46 pKa = 3.99IPAGSIYY53 pKa = 10.67SGEE56 pKa = 4.07TGIVSRR62 pKa = 11.84FAVEE66 pKa = 4.5LGAGSAAGEE75 pKa = 4.22TCGAILFHH83 pKa = 7.09PNDD86 pKa = 3.18NTLAVFTQANPATTFVVNAGNFVNTNGPGQLFLQSNAAKK125 pKa = 9.21MRR127 pKa = 11.84SLAACITAMPTSSTLNCTGDD147 pKa = 3.57VAVGNITLSSVYY159 pKa = 10.39GATVSINSIFSLLTVRR175 pKa = 11.84GPMVRR180 pKa = 11.84KK181 pKa = 9.95NYY183 pKa = 7.54EE184 pKa = 4.0CKK186 pKa = 9.56MVPGAFDD193 pKa = 2.98SRR195 pKa = 11.84YY196 pKa = 10.16AKK198 pKa = 10.5VIAAGNPSTTGTDD211 pKa = 3.52DD212 pKa = 3.66SDD214 pKa = 3.71TSVICLAFRR223 pKa = 11.84GLPAASGLLYY233 pKa = 10.73RR234 pKa = 11.84LTSVVEE240 pKa = 3.59WTPILNVGLTATTTSNVGIKK260 pKa = 10.08HH261 pKa = 5.28EE262 pKa = 4.26QVVSALSKK270 pKa = 10.64SHH272 pKa = 6.29GGWWHH277 pKa = 6.05NLWNNIGSDD286 pKa = 3.47LGGVASHH293 pKa = 6.75LTSQVLTGGAKK304 pKa = 10.26AIAGLLLL311 pKa = 3.99
MM1 pKa = 7.59AKK3 pKa = 9.98KK4 pKa = 10.29SNKK7 pKa = 9.39SPQVQKK13 pKa = 6.72TTRR16 pKa = 11.84PKK18 pKa = 9.0RR19 pKa = 11.84QRR21 pKa = 11.84KK22 pKa = 8.41GRR24 pKa = 11.84IAGVQGKK31 pKa = 9.12ALQLARR37 pKa = 11.84LLNDD41 pKa = 3.58PCSAEE46 pKa = 3.99IPAGSIYY53 pKa = 10.67SGEE56 pKa = 4.07TGIVSRR62 pKa = 11.84FAVEE66 pKa = 4.5LGAGSAAGEE75 pKa = 4.22TCGAILFHH83 pKa = 7.09PNDD86 pKa = 3.18NTLAVFTQANPATTFVVNAGNFVNTNGPGQLFLQSNAAKK125 pKa = 9.21MRR127 pKa = 11.84SLAACITAMPTSSTLNCTGDD147 pKa = 3.57VAVGNITLSSVYY159 pKa = 10.39GATVSINSIFSLLTVRR175 pKa = 11.84GPMVRR180 pKa = 11.84KK181 pKa = 9.95NYY183 pKa = 7.54EE184 pKa = 4.0CKK186 pKa = 9.56MVPGAFDD193 pKa = 2.98SRR195 pKa = 11.84YY196 pKa = 10.16AKK198 pKa = 10.5VIAAGNPSTTGTDD211 pKa = 3.52DD212 pKa = 3.66SDD214 pKa = 3.71TSVICLAFRR223 pKa = 11.84GLPAASGLLYY233 pKa = 10.73RR234 pKa = 11.84LTSVVEE240 pKa = 3.59WTPILNVGLTATTTSNVGIKK260 pKa = 10.08HH261 pKa = 5.28EE262 pKa = 4.26QVVSALSKK270 pKa = 10.64SHH272 pKa = 6.29GGWWHH277 pKa = 6.05NLWNNIGSDD286 pKa = 3.47LGGVASHH293 pKa = 6.75LTSQVLTGGAKK304 pKa = 10.26AIAGLLLL311 pKa = 3.99
Molecular weight: 32.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
828 |
311 |
517 |
414.0 |
45.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.213 ± 1.717 | 2.415 ± 0.274 |
4.106 ± 0.866 | 5.314 ± 1.729 |
4.348 ± 0.821 | 8.454 ± 1.036 |
3.019 ± 0.797 | 4.348 ± 0.268 |
5.314 ± 0.459 | 9.662 ± 0.19 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.295 ± 0.388 | 4.589 ± 1.04 |
5.314 ± 0.64 | 2.899 ± 0.179 |
5.676 ± 1.026 | 7.246 ± 1.173 |
6.039 ± 1.674 | 6.643 ± 0.788 |
1.57 ± 0.16 | 2.536 ± 0.524 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
