
Hubei tombus-like virus 36
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
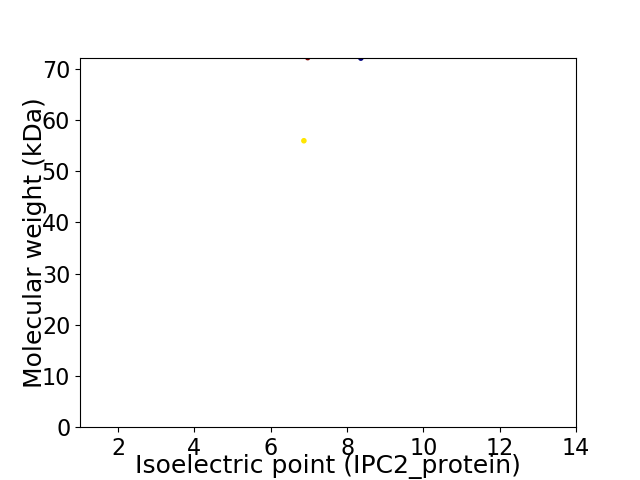
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGD7|A0A1L3KGD7_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 36 OX=1923284 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84SGVRR6 pKa = 11.84KK7 pKa = 9.93RR8 pKa = 11.84FAIKK12 pKa = 9.69TPTPKK17 pKa = 9.93PGKK20 pKa = 9.67LDD22 pKa = 3.49RR23 pKa = 11.84LGEE26 pKa = 4.26FVDD29 pKa = 5.03KK30 pKa = 10.45WLQQNLTPLAPDD42 pKa = 3.1SDD44 pKa = 4.15TTVEE48 pKa = 3.93HH49 pKa = 6.85WLSQTNYY56 pKa = 7.67PAYY59 pKa = 10.3RR60 pKa = 11.84RR61 pKa = 11.84AEE63 pKa = 4.03LLDD66 pKa = 4.11KK67 pKa = 10.91NDD69 pKa = 4.99KK70 pKa = 8.75ITNDD74 pKa = 3.11MDD76 pKa = 3.83TKK78 pKa = 10.82LFRR81 pKa = 11.84VKK83 pKa = 10.78SFMKK87 pKa = 10.3DD88 pKa = 2.72EE89 pKa = 5.02CYY91 pKa = 10.28PEE93 pKa = 4.21YY94 pKa = 10.96KK95 pKa = 9.73HH96 pKa = 6.74ARR98 pKa = 11.84AINSRR103 pKa = 11.84TDD105 pKa = 3.16EE106 pKa = 4.41FKK108 pKa = 10.66TLVGPTFKK116 pKa = 10.87LIEE119 pKa = 4.5KK120 pKa = 10.03EE121 pKa = 4.14LFEE124 pKa = 5.27LPWFIKK130 pKa = 10.1KK131 pKa = 10.04IPVKK135 pKa = 10.53DD136 pKa = 3.43RR137 pKa = 11.84PNYY140 pKa = 9.56IMEE143 pKa = 4.49RR144 pKa = 11.84LYY146 pKa = 10.94RR147 pKa = 11.84PNGKK151 pKa = 10.01YY152 pKa = 9.97YY153 pKa = 9.98ATDD156 pKa = 3.34YY157 pKa = 10.34TAFEE161 pKa = 3.97AHH163 pKa = 5.35FTKK166 pKa = 10.46PVMDD170 pKa = 3.81ACEE173 pKa = 3.9NRR175 pKa = 11.84LYY177 pKa = 10.91KK178 pKa = 10.96YY179 pKa = 7.57MTQHH183 pKa = 6.87LPNHH187 pKa = 6.48KK188 pKa = 10.3KK189 pKa = 10.42FMWYY193 pKa = 8.73IDD195 pKa = 3.57NVIGGRR201 pKa = 11.84NVCDD205 pKa = 4.99FKK207 pKa = 11.42HH208 pKa = 5.46LTVEE212 pKa = 3.95IDD214 pKa = 3.23ATRR217 pKa = 11.84MSGEE221 pKa = 4.25MNTSLGNGFSNLMLFLFMAHH241 pKa = 6.88EE242 pKa = 4.72KK243 pKa = 10.93GCTNVTGVVEE253 pKa = 4.37GDD255 pKa = 3.58DD256 pKa = 3.64GLFVLEE262 pKa = 4.74GPGITEE268 pKa = 4.06EE269 pKa = 4.14DD270 pKa = 3.32AKK272 pKa = 11.3EE273 pKa = 3.87CGFFLKK279 pKa = 10.22IEE281 pKa = 4.17SHH283 pKa = 6.58LSIATASFCGIIFDD297 pKa = 4.72EE298 pKa = 4.26EE299 pKa = 4.12DD300 pKa = 3.67RR301 pKa = 11.84NNLTNPLAEE310 pKa = 4.18LVNFGWTTGQYY321 pKa = 10.66ARR323 pKa = 11.84SKK325 pKa = 9.83RR326 pKa = 11.84ARR328 pKa = 11.84HH329 pKa = 5.75LEE331 pKa = 3.86LLRR334 pKa = 11.84SKK336 pKa = 10.73ALSLFYY342 pKa = 10.13QYY344 pKa = 10.26PGCPVLSSLANYY356 pKa = 10.23ALRR359 pKa = 11.84CTIGYY364 pKa = 8.15KK365 pKa = 10.52AKK367 pKa = 10.8LKK369 pKa = 10.18FFRR372 pKa = 11.84NMWEE376 pKa = 3.61RR377 pKa = 11.84EE378 pKa = 3.9QFIEE382 pKa = 3.97ALKK385 pKa = 10.8FKK387 pKa = 10.76DD388 pKa = 3.55EE389 pKa = 4.91AIVRR393 pKa = 11.84PVGNGSRR400 pKa = 11.84LLVEE404 pKa = 4.39EE405 pKa = 4.61LFGLSVTDD413 pKa = 3.45QLRR416 pKa = 11.84TEE418 pKa = 4.88AYY420 pKa = 10.3LDD422 pKa = 3.65SLDD425 pKa = 4.47TIQEE429 pKa = 4.05LDD431 pKa = 3.94MPWLEE436 pKa = 4.34WNYY439 pKa = 9.72HH440 pKa = 4.74KK441 pKa = 10.25HH442 pKa = 5.15WKK444 pKa = 9.96HH445 pKa = 5.62YY446 pKa = 9.89ADD448 pKa = 4.51NYY450 pKa = 10.13LAYY453 pKa = 9.74TDD455 pKa = 4.24GRR457 pKa = 11.84FKK459 pKa = 11.09ADD461 pKa = 3.13YY462 pKa = 10.13KK463 pKa = 10.56EE464 pKa = 3.89RR465 pKa = 11.84HH466 pKa = 5.8EE467 pKa = 4.19MSIHH471 pKa = 5.47SWTFTRR477 pKa = 11.84ACC479 pKa = 4.11
MM1 pKa = 7.75RR2 pKa = 11.84SGVRR6 pKa = 11.84KK7 pKa = 9.93RR8 pKa = 11.84FAIKK12 pKa = 9.69TPTPKK17 pKa = 9.93PGKK20 pKa = 9.67LDD22 pKa = 3.49RR23 pKa = 11.84LGEE26 pKa = 4.26FVDD29 pKa = 5.03KK30 pKa = 10.45WLQQNLTPLAPDD42 pKa = 3.1SDD44 pKa = 4.15TTVEE48 pKa = 3.93HH49 pKa = 6.85WLSQTNYY56 pKa = 7.67PAYY59 pKa = 10.3RR60 pKa = 11.84RR61 pKa = 11.84AEE63 pKa = 4.03LLDD66 pKa = 4.11KK67 pKa = 10.91NDD69 pKa = 4.99KK70 pKa = 8.75ITNDD74 pKa = 3.11MDD76 pKa = 3.83TKK78 pKa = 10.82LFRR81 pKa = 11.84VKK83 pKa = 10.78SFMKK87 pKa = 10.3DD88 pKa = 2.72EE89 pKa = 5.02CYY91 pKa = 10.28PEE93 pKa = 4.21YY94 pKa = 10.96KK95 pKa = 9.73HH96 pKa = 6.74ARR98 pKa = 11.84AINSRR103 pKa = 11.84TDD105 pKa = 3.16EE106 pKa = 4.41FKK108 pKa = 10.66TLVGPTFKK116 pKa = 10.87LIEE119 pKa = 4.5KK120 pKa = 10.03EE121 pKa = 4.14LFEE124 pKa = 5.27LPWFIKK130 pKa = 10.1KK131 pKa = 10.04IPVKK135 pKa = 10.53DD136 pKa = 3.43RR137 pKa = 11.84PNYY140 pKa = 9.56IMEE143 pKa = 4.49RR144 pKa = 11.84LYY146 pKa = 10.94RR147 pKa = 11.84PNGKK151 pKa = 10.01YY152 pKa = 9.97YY153 pKa = 9.98ATDD156 pKa = 3.34YY157 pKa = 10.34TAFEE161 pKa = 3.97AHH163 pKa = 5.35FTKK166 pKa = 10.46PVMDD170 pKa = 3.81ACEE173 pKa = 3.9NRR175 pKa = 11.84LYY177 pKa = 10.91KK178 pKa = 10.96YY179 pKa = 7.57MTQHH183 pKa = 6.87LPNHH187 pKa = 6.48KK188 pKa = 10.3KK189 pKa = 10.42FMWYY193 pKa = 8.73IDD195 pKa = 3.57NVIGGRR201 pKa = 11.84NVCDD205 pKa = 4.99FKK207 pKa = 11.42HH208 pKa = 5.46LTVEE212 pKa = 3.95IDD214 pKa = 3.23ATRR217 pKa = 11.84MSGEE221 pKa = 4.25MNTSLGNGFSNLMLFLFMAHH241 pKa = 6.88EE242 pKa = 4.72KK243 pKa = 10.93GCTNVTGVVEE253 pKa = 4.37GDD255 pKa = 3.58DD256 pKa = 3.64GLFVLEE262 pKa = 4.74GPGITEE268 pKa = 4.06EE269 pKa = 4.14DD270 pKa = 3.32AKK272 pKa = 11.3EE273 pKa = 3.87CGFFLKK279 pKa = 10.22IEE281 pKa = 4.17SHH283 pKa = 6.58LSIATASFCGIIFDD297 pKa = 4.72EE298 pKa = 4.26EE299 pKa = 4.12DD300 pKa = 3.67RR301 pKa = 11.84NNLTNPLAEE310 pKa = 4.18LVNFGWTTGQYY321 pKa = 10.66ARR323 pKa = 11.84SKK325 pKa = 9.83RR326 pKa = 11.84ARR328 pKa = 11.84HH329 pKa = 5.75LEE331 pKa = 3.86LLRR334 pKa = 11.84SKK336 pKa = 10.73ALSLFYY342 pKa = 10.13QYY344 pKa = 10.26PGCPVLSSLANYY356 pKa = 10.23ALRR359 pKa = 11.84CTIGYY364 pKa = 8.15KK365 pKa = 10.52AKK367 pKa = 10.8LKK369 pKa = 10.18FFRR372 pKa = 11.84NMWEE376 pKa = 3.61RR377 pKa = 11.84EE378 pKa = 3.9QFIEE382 pKa = 3.97ALKK385 pKa = 10.8FKK387 pKa = 10.76DD388 pKa = 3.55EE389 pKa = 4.91AIVRR393 pKa = 11.84PVGNGSRR400 pKa = 11.84LLVEE404 pKa = 4.39EE405 pKa = 4.61LFGLSVTDD413 pKa = 3.45QLRR416 pKa = 11.84TEE418 pKa = 4.88AYY420 pKa = 10.3LDD422 pKa = 3.65SLDD425 pKa = 4.47TIQEE429 pKa = 4.05LDD431 pKa = 3.94MPWLEE436 pKa = 4.34WNYY439 pKa = 9.72HH440 pKa = 4.74KK441 pKa = 10.25HH442 pKa = 5.15WKK444 pKa = 9.96HH445 pKa = 5.62YY446 pKa = 9.89ADD448 pKa = 4.51NYY450 pKa = 10.13LAYY453 pKa = 9.74TDD455 pKa = 4.24GRR457 pKa = 11.84FKK459 pKa = 11.09ADD461 pKa = 3.13YY462 pKa = 10.13KK463 pKa = 10.56EE464 pKa = 3.89RR465 pKa = 11.84HH466 pKa = 5.8EE467 pKa = 4.19MSIHH471 pKa = 5.47SWTFTRR477 pKa = 11.84ACC479 pKa = 4.11
Molecular weight: 55.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGD7|A0A1L3KGD7_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 36 OX=1923284 PE=4 SV=1
MM1 pKa = 7.65NDD3 pKa = 2.65NHH5 pKa = 6.94SIKK8 pKa = 10.53RR9 pKa = 11.84PEE11 pKa = 3.92KK12 pKa = 9.21EE13 pKa = 3.74GARR16 pKa = 11.84GGNKK20 pKa = 8.72KK21 pKa = 10.3RR22 pKa = 11.84KK23 pKa = 9.2GDD25 pKa = 3.95PRR27 pKa = 11.84PKK29 pKa = 9.62IWVEE33 pKa = 3.83KK34 pKa = 9.85KK35 pKa = 10.86GNGAGGGQPKK45 pKa = 9.96RR46 pKa = 11.84RR47 pKa = 11.84EE48 pKa = 3.82EE49 pKa = 4.27SVRR52 pKa = 11.84DD53 pKa = 3.54AAWYY57 pKa = 7.54TEE59 pKa = 4.1KK60 pKa = 10.73NAKK63 pKa = 8.46LTFLLNEE70 pKa = 4.35GSISEE75 pKa = 3.95PRR77 pKa = 11.84KK78 pKa = 9.94RR79 pKa = 11.84EE80 pKa = 3.54LSAQYY85 pKa = 7.45EE86 pKa = 4.45TNLYY90 pKa = 9.24EE91 pKa = 4.32LHH93 pKa = 7.04KK94 pKa = 10.45LKK96 pKa = 10.44QRR98 pKa = 11.84NAMVQEE104 pKa = 3.71PRR106 pKa = 11.84MPDD109 pKa = 3.32KK110 pKa = 10.92QPEE113 pKa = 4.33LPPPAVARR121 pKa = 11.84SVEE124 pKa = 4.22PSAKK128 pKa = 8.89TAARR132 pKa = 11.84KK133 pKa = 10.1DD134 pKa = 3.17KK135 pKa = 11.07GGPRR139 pKa = 11.84KK140 pKa = 9.88QGTRR144 pKa = 11.84AATRR148 pKa = 11.84KK149 pKa = 9.78SAVEE153 pKa = 3.55KK154 pKa = 11.06SLINSEE160 pKa = 4.33AKK162 pKa = 10.65AKK164 pKa = 10.9GIDD167 pKa = 3.64DD168 pKa = 3.77ARR170 pKa = 11.84QDD172 pKa = 3.28RR173 pKa = 11.84VEE175 pKa = 4.29FARR178 pKa = 11.84QDD180 pKa = 3.58VQSANAILDD189 pKa = 3.65NLLQQQAAPKK199 pKa = 9.93PEE201 pKa = 4.4EE202 pKa = 4.12EE203 pKa = 5.45DD204 pKa = 3.27EE205 pKa = 4.31DD206 pKa = 4.0TSEE209 pKa = 4.39EE210 pKa = 4.2EE211 pKa = 4.08EE212 pKa = 4.45SVEE215 pKa = 3.97PKK217 pKa = 10.77APEE220 pKa = 3.78TTADD224 pKa = 3.07KK225 pKa = 10.32WLSDD229 pKa = 4.59LYY231 pKa = 10.93RR232 pKa = 11.84CDD234 pKa = 2.99WLGPHH239 pKa = 6.62SKK241 pKa = 9.01FTFWEE246 pKa = 4.49GRR248 pKa = 11.84AYY250 pKa = 10.58HH251 pKa = 6.41LTTIAARR258 pKa = 11.84LILATLLWLVLCSFICNSLQGIAYY282 pKa = 7.14TYY284 pKa = 10.98LSAHH288 pKa = 6.77DD289 pKa = 4.65ADD291 pKa = 4.05LTRR294 pKa = 11.84AKK296 pKa = 10.59ALEE299 pKa = 4.35CFVGKK304 pKa = 10.35YY305 pKa = 9.81DD306 pKa = 3.93QGRR309 pKa = 11.84SYY311 pKa = 8.27WTLGGGLTKK320 pKa = 10.52LDD322 pKa = 4.0VKK324 pKa = 10.89PGDD327 pKa = 3.47PYY329 pKa = 11.47YY330 pKa = 11.29DD331 pKa = 3.41HH332 pKa = 7.57FYY334 pKa = 10.87DD335 pKa = 4.61SSGSAIKK342 pKa = 10.12GAKK345 pKa = 9.64NIPTQRR351 pKa = 11.84IMEE354 pKa = 4.82SIRR357 pKa = 11.84KK358 pKa = 8.64LRR360 pKa = 11.84QSSANALTAYY370 pKa = 10.38NWISNLSTFLYY381 pKa = 8.63QGKK384 pKa = 9.25YY385 pKa = 7.24LTGEE389 pKa = 3.74AFFFIILFFLWGKK402 pKa = 9.79IYY404 pKa = 10.72HH405 pKa = 6.6SDD407 pKa = 3.62TVKK410 pKa = 10.64HH411 pKa = 6.25SYY413 pKa = 11.12HH414 pKa = 5.55NVEE417 pKa = 4.27YY418 pKa = 10.28FDD420 pKa = 4.03PKK422 pKa = 9.13TWRR425 pKa = 11.84ACDD428 pKa = 3.43KK429 pKa = 10.15DD430 pKa = 3.87APGAVTADD438 pKa = 3.69YY439 pKa = 10.84FNTARR444 pKa = 11.84DD445 pKa = 3.47MRR447 pKa = 11.84NDD449 pKa = 2.76VSARR453 pKa = 11.84SKK455 pKa = 10.92KK456 pKa = 7.93EE457 pKa = 3.6HH458 pKa = 6.28EE459 pKa = 4.34PLYY462 pKa = 10.2FAYY465 pKa = 9.35TRR467 pKa = 11.84HH468 pKa = 5.99EE469 pKa = 4.15EE470 pKa = 3.9HH471 pKa = 5.98WWYY474 pKa = 10.37IGVGRR479 pKa = 11.84MKK481 pKa = 10.68VPVFRR486 pKa = 11.84HH487 pKa = 4.92IKK489 pKa = 9.75KK490 pKa = 10.07VPNVKK495 pKa = 10.03LSSMEE500 pKa = 4.21LVFQICHH507 pKa = 6.44PFNMEE512 pKa = 3.02WTAPEE517 pKa = 5.07DD518 pKa = 3.35ICIDD522 pKa = 3.76RR523 pKa = 11.84MTKK526 pKa = 8.92TKK528 pKa = 9.68NTTQSININKK538 pKa = 9.0QLIITKK544 pKa = 9.88QDD546 pKa = 2.86VFEE549 pKa = 4.19NSRR552 pKa = 11.84AISLGFFYY560 pKa = 10.9GLRR563 pKa = 11.84QDD565 pKa = 3.56HH566 pKa = 6.66SKK568 pKa = 10.92SAFQLAPEE576 pKa = 4.95LKK578 pKa = 10.22AAYY581 pKa = 10.04QFDD584 pKa = 4.19FLHH587 pKa = 6.76TDD589 pKa = 3.36TVDD592 pKa = 3.45TRR594 pKa = 11.84CLYY597 pKa = 11.35LMTQISGLARR607 pKa = 11.84AYY609 pKa = 10.21RR610 pKa = 11.84SYY612 pKa = 11.61VHH614 pKa = 6.6MGQTLVSLCLFLWAYY629 pKa = 7.43MM630 pKa = 3.96
MM1 pKa = 7.65NDD3 pKa = 2.65NHH5 pKa = 6.94SIKK8 pKa = 10.53RR9 pKa = 11.84PEE11 pKa = 3.92KK12 pKa = 9.21EE13 pKa = 3.74GARR16 pKa = 11.84GGNKK20 pKa = 8.72KK21 pKa = 10.3RR22 pKa = 11.84KK23 pKa = 9.2GDD25 pKa = 3.95PRR27 pKa = 11.84PKK29 pKa = 9.62IWVEE33 pKa = 3.83KK34 pKa = 9.85KK35 pKa = 10.86GNGAGGGQPKK45 pKa = 9.96RR46 pKa = 11.84RR47 pKa = 11.84EE48 pKa = 3.82EE49 pKa = 4.27SVRR52 pKa = 11.84DD53 pKa = 3.54AAWYY57 pKa = 7.54TEE59 pKa = 4.1KK60 pKa = 10.73NAKK63 pKa = 8.46LTFLLNEE70 pKa = 4.35GSISEE75 pKa = 3.95PRR77 pKa = 11.84KK78 pKa = 9.94RR79 pKa = 11.84EE80 pKa = 3.54LSAQYY85 pKa = 7.45EE86 pKa = 4.45TNLYY90 pKa = 9.24EE91 pKa = 4.32LHH93 pKa = 7.04KK94 pKa = 10.45LKK96 pKa = 10.44QRR98 pKa = 11.84NAMVQEE104 pKa = 3.71PRR106 pKa = 11.84MPDD109 pKa = 3.32KK110 pKa = 10.92QPEE113 pKa = 4.33LPPPAVARR121 pKa = 11.84SVEE124 pKa = 4.22PSAKK128 pKa = 8.89TAARR132 pKa = 11.84KK133 pKa = 10.1DD134 pKa = 3.17KK135 pKa = 11.07GGPRR139 pKa = 11.84KK140 pKa = 9.88QGTRR144 pKa = 11.84AATRR148 pKa = 11.84KK149 pKa = 9.78SAVEE153 pKa = 3.55KK154 pKa = 11.06SLINSEE160 pKa = 4.33AKK162 pKa = 10.65AKK164 pKa = 10.9GIDD167 pKa = 3.64DD168 pKa = 3.77ARR170 pKa = 11.84QDD172 pKa = 3.28RR173 pKa = 11.84VEE175 pKa = 4.29FARR178 pKa = 11.84QDD180 pKa = 3.58VQSANAILDD189 pKa = 3.65NLLQQQAAPKK199 pKa = 9.93PEE201 pKa = 4.4EE202 pKa = 4.12EE203 pKa = 5.45DD204 pKa = 3.27EE205 pKa = 4.31DD206 pKa = 4.0TSEE209 pKa = 4.39EE210 pKa = 4.2EE211 pKa = 4.08EE212 pKa = 4.45SVEE215 pKa = 3.97PKK217 pKa = 10.77APEE220 pKa = 3.78TTADD224 pKa = 3.07KK225 pKa = 10.32WLSDD229 pKa = 4.59LYY231 pKa = 10.93RR232 pKa = 11.84CDD234 pKa = 2.99WLGPHH239 pKa = 6.62SKK241 pKa = 9.01FTFWEE246 pKa = 4.49GRR248 pKa = 11.84AYY250 pKa = 10.58HH251 pKa = 6.41LTTIAARR258 pKa = 11.84LILATLLWLVLCSFICNSLQGIAYY282 pKa = 7.14TYY284 pKa = 10.98LSAHH288 pKa = 6.77DD289 pKa = 4.65ADD291 pKa = 4.05LTRR294 pKa = 11.84AKK296 pKa = 10.59ALEE299 pKa = 4.35CFVGKK304 pKa = 10.35YY305 pKa = 9.81DD306 pKa = 3.93QGRR309 pKa = 11.84SYY311 pKa = 8.27WTLGGGLTKK320 pKa = 10.52LDD322 pKa = 4.0VKK324 pKa = 10.89PGDD327 pKa = 3.47PYY329 pKa = 11.47YY330 pKa = 11.29DD331 pKa = 3.41HH332 pKa = 7.57FYY334 pKa = 10.87DD335 pKa = 4.61SSGSAIKK342 pKa = 10.12GAKK345 pKa = 9.64NIPTQRR351 pKa = 11.84IMEE354 pKa = 4.82SIRR357 pKa = 11.84KK358 pKa = 8.64LRR360 pKa = 11.84QSSANALTAYY370 pKa = 10.38NWISNLSTFLYY381 pKa = 8.63QGKK384 pKa = 9.25YY385 pKa = 7.24LTGEE389 pKa = 3.74AFFFIILFFLWGKK402 pKa = 9.79IYY404 pKa = 10.72HH405 pKa = 6.6SDD407 pKa = 3.62TVKK410 pKa = 10.64HH411 pKa = 6.25SYY413 pKa = 11.12HH414 pKa = 5.55NVEE417 pKa = 4.27YY418 pKa = 10.28FDD420 pKa = 4.03PKK422 pKa = 9.13TWRR425 pKa = 11.84ACDD428 pKa = 3.43KK429 pKa = 10.15DD430 pKa = 3.87APGAVTADD438 pKa = 3.69YY439 pKa = 10.84FNTARR444 pKa = 11.84DD445 pKa = 3.47MRR447 pKa = 11.84NDD449 pKa = 2.76VSARR453 pKa = 11.84SKK455 pKa = 10.92KK456 pKa = 7.93EE457 pKa = 3.6HH458 pKa = 6.28EE459 pKa = 4.34PLYY462 pKa = 10.2FAYY465 pKa = 9.35TRR467 pKa = 11.84HH468 pKa = 5.99EE469 pKa = 4.15EE470 pKa = 3.9HH471 pKa = 5.98WWYY474 pKa = 10.37IGVGRR479 pKa = 11.84MKK481 pKa = 10.68VPVFRR486 pKa = 11.84HH487 pKa = 4.92IKK489 pKa = 9.75KK490 pKa = 10.07VPNVKK495 pKa = 10.03LSSMEE500 pKa = 4.21LVFQICHH507 pKa = 6.44PFNMEE512 pKa = 3.02WTAPEE517 pKa = 5.07DD518 pKa = 3.35ICIDD522 pKa = 3.76RR523 pKa = 11.84MTKK526 pKa = 8.92TKK528 pKa = 9.68NTTQSININKK538 pKa = 9.0QLIITKK544 pKa = 9.88QDD546 pKa = 2.86VFEE549 pKa = 4.19NSRR552 pKa = 11.84AISLGFFYY560 pKa = 10.9GLRR563 pKa = 11.84QDD565 pKa = 3.56HH566 pKa = 6.66SKK568 pKa = 10.92SAFQLAPEE576 pKa = 4.95LKK578 pKa = 10.22AAYY581 pKa = 10.04QFDD584 pKa = 4.19FLHH587 pKa = 6.76TDD589 pKa = 3.36TVDD592 pKa = 3.45TRR594 pKa = 11.84CLYY597 pKa = 11.35LMTQISGLARR607 pKa = 11.84AYY609 pKa = 10.21RR610 pKa = 11.84SYY612 pKa = 11.61VHH614 pKa = 6.6MGQTLVSLCLFLWAYY629 pKa = 7.43MM630 pKa = 3.96
Molecular weight: 72.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1109 |
479 |
630 |
554.5 |
64.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.665 ± 0.888 | 1.623 ± 0.141 |
5.861 ± 0.107 | 6.943 ± 0.316 |
4.959 ± 0.604 | 5.5 ± 0.04 |
2.795 ± 0.07 | 4.418 ± 0.134 |
7.935 ± 0.116 | 9.017 ± 0.554 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.319 | 4.418 ± 0.327 |
4.418 ± 0.134 | 3.156 ± 0.704 |
6.222 ± 0.092 | 5.591 ± 0.665 |
6.222 ± 0.253 | 4.148 ± 0.015 |
2.164 ± 0.042 | 4.599 ± 0.112 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
