
Phenylobacterium deserti
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
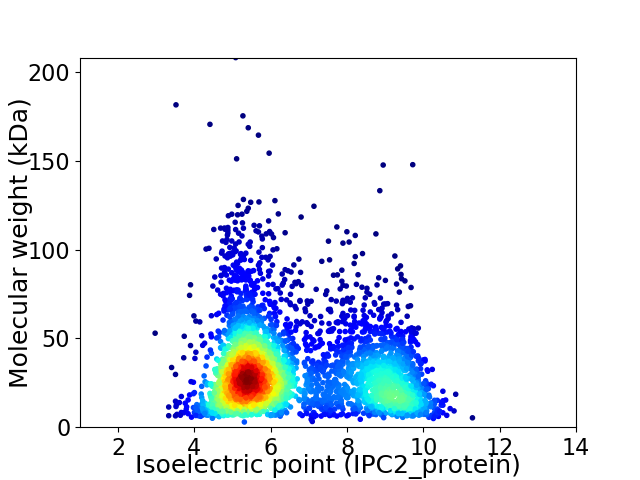
Virtual 2D-PAGE plot for 3698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
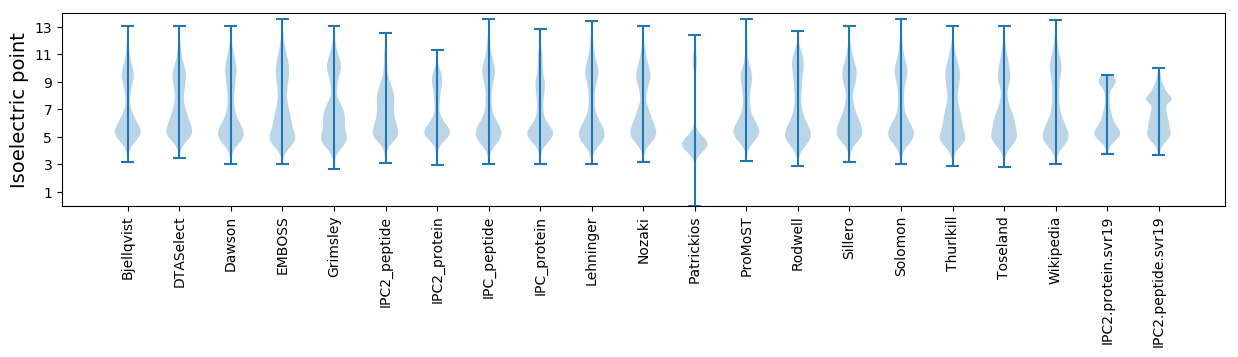
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328AV49|A0A328AV49_9CAUL Uncharacterized protein OS=Phenylobacterium deserti OX=1914756 GN=DJ018_02150 PE=4 SV=1
MM1 pKa = 7.12FASAHH6 pKa = 5.83NGAGLALNIARR17 pKa = 11.84EE18 pKa = 4.29EE19 pKa = 4.24TNYY22 pKa = 9.21MRR24 pKa = 11.84PNSMDD29 pKa = 3.1DD30 pKa = 3.24FARR33 pKa = 11.84ITRR36 pKa = 11.84GTRR39 pKa = 11.84DD40 pKa = 3.37AQKK43 pKa = 10.92PEE45 pKa = 3.89ATLSGLTPTSATFTHH60 pKa = 7.26ADD62 pKa = 3.27GAAPATTVGSLGTEE76 pKa = 4.01RR77 pKa = 11.84YY78 pKa = 10.13DD79 pKa = 4.7LIPVALTAGTTYY91 pKa = 11.03SFAYY95 pKa = 10.14RR96 pKa = 11.84PTEE99 pKa = 3.67TGGIEE104 pKa = 4.42DD105 pKa = 4.48PFLGLVNPAGTQFLAQDD122 pKa = 3.98DD123 pKa = 4.37DD124 pKa = 4.72GGLGRR129 pKa = 11.84SSMITFTPTASGTYY143 pKa = 10.55LLYY146 pKa = 7.97TTSWYY151 pKa = 10.52HH152 pKa = 6.64IDD154 pKa = 3.76PSAPGYY160 pKa = 9.76PNYY163 pKa = 9.98RR164 pKa = 11.84DD165 pKa = 3.31AGGYY169 pKa = 7.2TVDD172 pKa = 3.85MWTADD177 pKa = 3.85PATDD181 pKa = 4.07APSTRR186 pKa = 11.84AGALTIDD193 pKa = 3.08VGTTYY198 pKa = 11.07GHH200 pKa = 7.46LDD202 pKa = 3.22AAGDD206 pKa = 3.38QDD208 pKa = 3.93MYY210 pKa = 10.85RR211 pKa = 11.84VEE213 pKa = 4.7LDD215 pKa = 2.99PGLFYY220 pKa = 10.86TFTYY224 pKa = 10.39AGGIASSAEE233 pKa = 4.08YY234 pKa = 9.82PGEE237 pKa = 4.02LPGEE241 pKa = 4.44SIGVLRR247 pKa = 11.84LYY249 pKa = 10.44DD250 pKa = 3.58AEE252 pKa = 4.34GNEE255 pKa = 4.07LSAAVNYY262 pKa = 7.47EE263 pKa = 3.86TGLGFLSPEE272 pKa = 3.8GGTYY276 pKa = 8.44YY277 pKa = 10.79LRR279 pKa = 11.84VEE281 pKa = 4.63GYY283 pKa = 9.87EE284 pKa = 3.83PDD286 pKa = 3.29MTGGYY291 pKa = 7.72TLDD294 pKa = 4.36VKK296 pKa = 11.14AQDD299 pKa = 3.91PTDD302 pKa = 4.15FDD304 pKa = 4.49PLEE307 pKa = 4.19SLNWDD312 pKa = 3.52EE313 pKa = 5.39ADD315 pKa = 4.33NIPTTMVNGVPTAYY329 pKa = 10.51VYY331 pKa = 10.06FASARR336 pKa = 11.84EE337 pKa = 4.12GGFGEE342 pKa = 4.5TEE344 pKa = 4.23DD345 pKa = 5.76DD346 pKa = 3.98GVTPITTYY354 pKa = 10.46GWEE357 pKa = 3.82QFQIQGVMRR366 pKa = 11.84ALQEE370 pKa = 3.97YY371 pKa = 8.92TPITGINYY379 pKa = 8.71VRR381 pKa = 11.84TTDD384 pKa = 3.06INQATFRR391 pKa = 11.84LVTTINDD398 pKa = 3.48DD399 pKa = 3.28YY400 pKa = 11.5GARR403 pKa = 11.84FNPRR407 pKa = 11.84DD408 pKa = 3.63PSAGDD413 pKa = 3.44LQGVGVFNLASGGFTRR429 pKa = 11.84PEE431 pKa = 3.8TLEE434 pKa = 4.17PGGYY438 pKa = 9.13SYY440 pKa = 11.74AVVLHH445 pKa = 6.1EE446 pKa = 5.07FGHH449 pKa = 5.58AHH451 pKa = 6.27GVAHH455 pKa = 6.5PHH457 pKa = 6.86DD458 pKa = 4.48EE459 pKa = 4.64GGGSEE464 pKa = 4.12ILLGVTGPTGSLGIYY479 pKa = 9.9DD480 pKa = 4.96LNQGVYY486 pKa = 8.91TVMSYY491 pKa = 11.59NDD493 pKa = 3.39GWQTNPDD500 pKa = 3.38GPLEE504 pKa = 4.16FTRR507 pKa = 11.84QTWGSGWSGTLGAFDD522 pKa = 3.87IAVLQARR529 pKa = 11.84YY530 pKa = 9.16GVHH533 pKa = 6.14AHH535 pKa = 5.16NTGDD539 pKa = 3.4NVYY542 pKa = 11.03ALTGPQKK549 pKa = 10.42DD550 pKa = 4.03AYY552 pKa = 10.0YY553 pKa = 8.82QTIWDD558 pKa = 3.78TGGNDD563 pKa = 3.48TIAYY567 pKa = 9.86DD568 pKa = 3.97GGRR571 pKa = 11.84DD572 pKa = 3.41AHH574 pKa = 7.02IDD576 pKa = 3.65LLAATLDD583 pKa = 3.77YY584 pKa = 10.42TPTGGGVVSFVDD596 pKa = 4.0GTYY599 pKa = 10.93GGYY602 pKa = 9.01TIANGVVIEE611 pKa = 4.15NARR614 pKa = 11.84GGNGSDD620 pKa = 2.97ALMGNSVANRR630 pKa = 11.84LDD632 pKa = 3.58GQNGDD637 pKa = 4.02DD638 pKa = 3.8GLLGRR643 pKa = 11.84EE644 pKa = 4.4GDD646 pKa = 3.98DD647 pKa = 4.82LLIGGNGKK655 pKa = 8.44DD656 pKa = 3.62TLNGGTGNDD665 pKa = 3.86SLDD668 pKa = 3.56GGTGRR673 pKa = 11.84DD674 pKa = 3.77VLNGGAGDD682 pKa = 3.78DD683 pKa = 3.86TLNGGLHH690 pKa = 7.34DD691 pKa = 4.84DD692 pKa = 4.11LFVFEE697 pKa = 5.59DD698 pKa = 3.86AGTDD702 pKa = 3.69TIVGYY707 pKa = 10.23QKK709 pKa = 11.15GEE711 pKa = 4.03QFDD714 pKa = 4.56LSALNVTWADD724 pKa = 3.36VTIEE728 pKa = 4.0SGRR731 pKa = 11.84IGVDD735 pKa = 3.38LEE737 pKa = 4.2GDD739 pKa = 3.21QDD741 pKa = 3.61LTIIVNTSGITQGNFIFAA759 pKa = 4.52
MM1 pKa = 7.12FASAHH6 pKa = 5.83NGAGLALNIARR17 pKa = 11.84EE18 pKa = 4.29EE19 pKa = 4.24TNYY22 pKa = 9.21MRR24 pKa = 11.84PNSMDD29 pKa = 3.1DD30 pKa = 3.24FARR33 pKa = 11.84ITRR36 pKa = 11.84GTRR39 pKa = 11.84DD40 pKa = 3.37AQKK43 pKa = 10.92PEE45 pKa = 3.89ATLSGLTPTSATFTHH60 pKa = 7.26ADD62 pKa = 3.27GAAPATTVGSLGTEE76 pKa = 4.01RR77 pKa = 11.84YY78 pKa = 10.13DD79 pKa = 4.7LIPVALTAGTTYY91 pKa = 11.03SFAYY95 pKa = 10.14RR96 pKa = 11.84PTEE99 pKa = 3.67TGGIEE104 pKa = 4.42DD105 pKa = 4.48PFLGLVNPAGTQFLAQDD122 pKa = 3.98DD123 pKa = 4.37DD124 pKa = 4.72GGLGRR129 pKa = 11.84SSMITFTPTASGTYY143 pKa = 10.55LLYY146 pKa = 7.97TTSWYY151 pKa = 10.52HH152 pKa = 6.64IDD154 pKa = 3.76PSAPGYY160 pKa = 9.76PNYY163 pKa = 9.98RR164 pKa = 11.84DD165 pKa = 3.31AGGYY169 pKa = 7.2TVDD172 pKa = 3.85MWTADD177 pKa = 3.85PATDD181 pKa = 4.07APSTRR186 pKa = 11.84AGALTIDD193 pKa = 3.08VGTTYY198 pKa = 11.07GHH200 pKa = 7.46LDD202 pKa = 3.22AAGDD206 pKa = 3.38QDD208 pKa = 3.93MYY210 pKa = 10.85RR211 pKa = 11.84VEE213 pKa = 4.7LDD215 pKa = 2.99PGLFYY220 pKa = 10.86TFTYY224 pKa = 10.39AGGIASSAEE233 pKa = 4.08YY234 pKa = 9.82PGEE237 pKa = 4.02LPGEE241 pKa = 4.44SIGVLRR247 pKa = 11.84LYY249 pKa = 10.44DD250 pKa = 3.58AEE252 pKa = 4.34GNEE255 pKa = 4.07LSAAVNYY262 pKa = 7.47EE263 pKa = 3.86TGLGFLSPEE272 pKa = 3.8GGTYY276 pKa = 8.44YY277 pKa = 10.79LRR279 pKa = 11.84VEE281 pKa = 4.63GYY283 pKa = 9.87EE284 pKa = 3.83PDD286 pKa = 3.29MTGGYY291 pKa = 7.72TLDD294 pKa = 4.36VKK296 pKa = 11.14AQDD299 pKa = 3.91PTDD302 pKa = 4.15FDD304 pKa = 4.49PLEE307 pKa = 4.19SLNWDD312 pKa = 3.52EE313 pKa = 5.39ADD315 pKa = 4.33NIPTTMVNGVPTAYY329 pKa = 10.51VYY331 pKa = 10.06FASARR336 pKa = 11.84EE337 pKa = 4.12GGFGEE342 pKa = 4.5TEE344 pKa = 4.23DD345 pKa = 5.76DD346 pKa = 3.98GVTPITTYY354 pKa = 10.46GWEE357 pKa = 3.82QFQIQGVMRR366 pKa = 11.84ALQEE370 pKa = 3.97YY371 pKa = 8.92TPITGINYY379 pKa = 8.71VRR381 pKa = 11.84TTDD384 pKa = 3.06INQATFRR391 pKa = 11.84LVTTINDD398 pKa = 3.48DD399 pKa = 3.28YY400 pKa = 11.5GARR403 pKa = 11.84FNPRR407 pKa = 11.84DD408 pKa = 3.63PSAGDD413 pKa = 3.44LQGVGVFNLASGGFTRR429 pKa = 11.84PEE431 pKa = 3.8TLEE434 pKa = 4.17PGGYY438 pKa = 9.13SYY440 pKa = 11.74AVVLHH445 pKa = 6.1EE446 pKa = 5.07FGHH449 pKa = 5.58AHH451 pKa = 6.27GVAHH455 pKa = 6.5PHH457 pKa = 6.86DD458 pKa = 4.48EE459 pKa = 4.64GGGSEE464 pKa = 4.12ILLGVTGPTGSLGIYY479 pKa = 9.9DD480 pKa = 4.96LNQGVYY486 pKa = 8.91TVMSYY491 pKa = 11.59NDD493 pKa = 3.39GWQTNPDD500 pKa = 3.38GPLEE504 pKa = 4.16FTRR507 pKa = 11.84QTWGSGWSGTLGAFDD522 pKa = 3.87IAVLQARR529 pKa = 11.84YY530 pKa = 9.16GVHH533 pKa = 6.14AHH535 pKa = 5.16NTGDD539 pKa = 3.4NVYY542 pKa = 11.03ALTGPQKK549 pKa = 10.42DD550 pKa = 4.03AYY552 pKa = 10.0YY553 pKa = 8.82QTIWDD558 pKa = 3.78TGGNDD563 pKa = 3.48TIAYY567 pKa = 9.86DD568 pKa = 3.97GGRR571 pKa = 11.84DD572 pKa = 3.41AHH574 pKa = 7.02IDD576 pKa = 3.65LLAATLDD583 pKa = 3.77YY584 pKa = 10.42TPTGGGVVSFVDD596 pKa = 4.0GTYY599 pKa = 10.93GGYY602 pKa = 9.01TIANGVVIEE611 pKa = 4.15NARR614 pKa = 11.84GGNGSDD620 pKa = 2.97ALMGNSVANRR630 pKa = 11.84LDD632 pKa = 3.58GQNGDD637 pKa = 4.02DD638 pKa = 3.8GLLGRR643 pKa = 11.84EE644 pKa = 4.4GDD646 pKa = 3.98DD647 pKa = 4.82LLIGGNGKK655 pKa = 8.44DD656 pKa = 3.62TLNGGTGNDD665 pKa = 3.86SLDD668 pKa = 3.56GGTGRR673 pKa = 11.84DD674 pKa = 3.77VLNGGAGDD682 pKa = 3.78DD683 pKa = 3.86TLNGGLHH690 pKa = 7.34DD691 pKa = 4.84DD692 pKa = 4.11LFVFEE697 pKa = 5.59DD698 pKa = 3.86AGTDD702 pKa = 3.69TIVGYY707 pKa = 10.23QKK709 pKa = 11.15GEE711 pKa = 4.03QFDD714 pKa = 4.56LSALNVTWADD724 pKa = 3.36VTIEE728 pKa = 4.0SGRR731 pKa = 11.84IGVDD735 pKa = 3.38LEE737 pKa = 4.2GDD739 pKa = 3.21QDD741 pKa = 3.61LTIIVNTSGITQGNFIFAA759 pKa = 4.52
Molecular weight: 80.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328AU99|A0A328AU99_9CAUL Beta-ketoacyl-ACP reductase OS=Phenylobacterium deserti OX=1914756 GN=DJ018_08010 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTKK25 pKa = 10.02NGQKK29 pKa = 9.43ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1169876 |
27 |
1930 |
316.4 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.846 ± 0.065 | 0.722 ± 0.011 |
5.54 ± 0.028 | 5.902 ± 0.046 |
3.536 ± 0.025 | 8.937 ± 0.039 |
1.778 ± 0.02 | 4.071 ± 0.027 |
2.858 ± 0.03 | 10.116 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.019 | 2.367 ± 0.03 |
5.655 ± 0.035 | 3.482 ± 0.024 |
7.729 ± 0.046 | 5.133 ± 0.033 |
5.061 ± 0.033 | 7.58 ± 0.034 |
1.438 ± 0.018 | 2.104 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
