
Chloropicon primus
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chloropicophyceae; Chloropicales; Chloropicaceae;
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
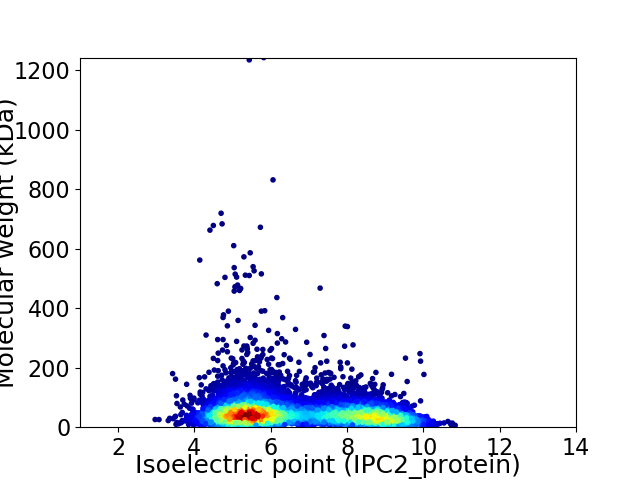
Virtual 2D-PAGE plot for 8606 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8MPD5|A0A5B8MPD5_9CHLO Transcription initiation factor IIF subunit beta OS=Chloropicon primus OX=1764295 GN=A3770_07p48870 PE=3 SV=1
MM1 pKa = 6.98VHH3 pKa = 5.69TSKK6 pKa = 11.25YY7 pKa = 9.76LAASLALALAVAGGAVAQDD26 pKa = 3.41QQDD29 pKa = 3.41FGDD32 pKa = 3.72AMRR35 pKa = 11.84QFLAHH40 pKa = 6.53SANGTCEE47 pKa = 5.51DD48 pKa = 4.38IPQEE52 pKa = 3.92MWNTTMRR59 pKa = 11.84NGTNQDD65 pKa = 3.59CEE67 pKa = 4.53GWWQSIAGDD76 pKa = 3.86ARR78 pKa = 11.84SVADD82 pKa = 5.22DD83 pKa = 3.26IGQAFEE89 pKa = 4.34NGAPLLVNGTEE100 pKa = 4.37KK101 pKa = 10.34ICVNSMPLDD110 pKa = 4.46DD111 pKa = 4.63YY112 pKa = 11.8SQIVQSACWPSGMEE126 pKa = 4.01EE127 pKa = 4.03EE128 pKa = 4.78PEE130 pKa = 3.89EE131 pKa = 3.96NHH133 pKa = 6.4AVRR136 pKa = 11.84IRR138 pKa = 11.84LGLPLQCVDD147 pKa = 3.75AADD150 pKa = 4.48YY151 pKa = 11.32DD152 pKa = 5.25DD153 pKa = 4.05STDD156 pKa = 3.15WSDD159 pKa = 4.15VPLQAYY165 pKa = 7.34MVSGEE170 pKa = 4.15VALCGDD176 pKa = 4.85DD177 pKa = 4.26AGEE180 pKa = 4.16DD181 pKa = 3.38QRR183 pKa = 11.84SYY185 pKa = 11.66VGLALGDD192 pKa = 3.9TIVAGDD198 pKa = 4.37NQTEE202 pKa = 4.68DD203 pKa = 4.02YY204 pKa = 11.38DD205 pKa = 5.89DD206 pKa = 6.01DD207 pKa = 6.06DD208 pKa = 6.98DD209 pKa = 6.02EE210 pKa = 6.81IEE212 pKa = 4.54DD213 pKa = 4.19HH214 pKa = 7.43DD215 pKa = 5.37GDD217 pKa = 3.84VDD219 pKa = 4.28GEE221 pKa = 4.28EE222 pKa = 4.24SQHH225 pKa = 7.7ADD227 pKa = 3.28FGDD230 pKa = 3.54MMKK233 pKa = 10.35QYY235 pKa = 11.11AKK237 pKa = 10.52DD238 pKa = 3.49AASEE242 pKa = 4.26TCEE245 pKa = 4.41PVPQEE250 pKa = 3.79LWNTTVRR257 pKa = 11.84DD258 pKa = 5.17GEE260 pKa = 4.25DD261 pKa = 3.39QDD263 pKa = 5.49CKK265 pKa = 11.13GWWQSIAGGARR276 pKa = 11.84SVADD280 pKa = 4.82DD281 pKa = 3.4IGQAFVGGVPLLMEE295 pKa = 5.78DD296 pKa = 3.81GTVADD301 pKa = 5.21CVTSLDD307 pKa = 3.07WDD309 pKa = 3.82YY310 pKa = 12.03DD311 pKa = 3.64FSQNVKK317 pKa = 9.34SACWPSGMEE326 pKa = 4.09EE327 pKa = 4.06MPEE330 pKa = 3.92GNHH333 pKa = 5.62AVRR336 pKa = 11.84VTLEE340 pKa = 4.48LPLKK344 pKa = 10.06CADD347 pKa = 3.55VNGPTMALADD357 pKa = 3.84IPEE360 pKa = 4.13EE361 pKa = 4.09TIVISGEE368 pKa = 4.03VAFCGEE374 pKa = 4.78DD375 pKa = 3.0AGEE378 pKa = 4.19DD379 pKa = 3.41QRR381 pKa = 11.84SYY383 pKa = 10.65RR384 pKa = 11.84WAAGDD389 pKa = 3.31QQ390 pKa = 3.47
MM1 pKa = 6.98VHH3 pKa = 5.69TSKK6 pKa = 11.25YY7 pKa = 9.76LAASLALALAVAGGAVAQDD26 pKa = 3.41QQDD29 pKa = 3.41FGDD32 pKa = 3.72AMRR35 pKa = 11.84QFLAHH40 pKa = 6.53SANGTCEE47 pKa = 5.51DD48 pKa = 4.38IPQEE52 pKa = 3.92MWNTTMRR59 pKa = 11.84NGTNQDD65 pKa = 3.59CEE67 pKa = 4.53GWWQSIAGDD76 pKa = 3.86ARR78 pKa = 11.84SVADD82 pKa = 5.22DD83 pKa = 3.26IGQAFEE89 pKa = 4.34NGAPLLVNGTEE100 pKa = 4.37KK101 pKa = 10.34ICVNSMPLDD110 pKa = 4.46DD111 pKa = 4.63YY112 pKa = 11.8SQIVQSACWPSGMEE126 pKa = 4.01EE127 pKa = 4.03EE128 pKa = 4.78PEE130 pKa = 3.89EE131 pKa = 3.96NHH133 pKa = 6.4AVRR136 pKa = 11.84IRR138 pKa = 11.84LGLPLQCVDD147 pKa = 3.75AADD150 pKa = 4.48YY151 pKa = 11.32DD152 pKa = 5.25DD153 pKa = 4.05STDD156 pKa = 3.15WSDD159 pKa = 4.15VPLQAYY165 pKa = 7.34MVSGEE170 pKa = 4.15VALCGDD176 pKa = 4.85DD177 pKa = 4.26AGEE180 pKa = 4.16DD181 pKa = 3.38QRR183 pKa = 11.84SYY185 pKa = 11.66VGLALGDD192 pKa = 3.9TIVAGDD198 pKa = 4.37NQTEE202 pKa = 4.68DD203 pKa = 4.02YY204 pKa = 11.38DD205 pKa = 5.89DD206 pKa = 6.01DD207 pKa = 6.06DD208 pKa = 6.98DD209 pKa = 6.02EE210 pKa = 6.81IEE212 pKa = 4.54DD213 pKa = 4.19HH214 pKa = 7.43DD215 pKa = 5.37GDD217 pKa = 3.84VDD219 pKa = 4.28GEE221 pKa = 4.28EE222 pKa = 4.24SQHH225 pKa = 7.7ADD227 pKa = 3.28FGDD230 pKa = 3.54MMKK233 pKa = 10.35QYY235 pKa = 11.11AKK237 pKa = 10.52DD238 pKa = 3.49AASEE242 pKa = 4.26TCEE245 pKa = 4.41PVPQEE250 pKa = 3.79LWNTTVRR257 pKa = 11.84DD258 pKa = 5.17GEE260 pKa = 4.25DD261 pKa = 3.39QDD263 pKa = 5.49CKK265 pKa = 11.13GWWQSIAGGARR276 pKa = 11.84SVADD280 pKa = 4.82DD281 pKa = 3.4IGQAFVGGVPLLMEE295 pKa = 5.78DD296 pKa = 3.81GTVADD301 pKa = 5.21CVTSLDD307 pKa = 3.07WDD309 pKa = 3.82YY310 pKa = 12.03DD311 pKa = 3.64FSQNVKK317 pKa = 9.34SACWPSGMEE326 pKa = 4.09EE327 pKa = 4.06MPEE330 pKa = 3.92GNHH333 pKa = 5.62AVRR336 pKa = 11.84VTLEE340 pKa = 4.48LPLKK344 pKa = 10.06CADD347 pKa = 3.55VNGPTMALADD357 pKa = 3.84IPEE360 pKa = 4.13EE361 pKa = 4.09TIVISGEE368 pKa = 4.03VAFCGEE374 pKa = 4.78DD375 pKa = 3.0AGEE378 pKa = 4.19DD379 pKa = 3.41QRR381 pKa = 11.84SYY383 pKa = 10.65RR384 pKa = 11.84WAAGDD389 pKa = 3.31QQ390 pKa = 3.47
Molecular weight: 42.16 kDa
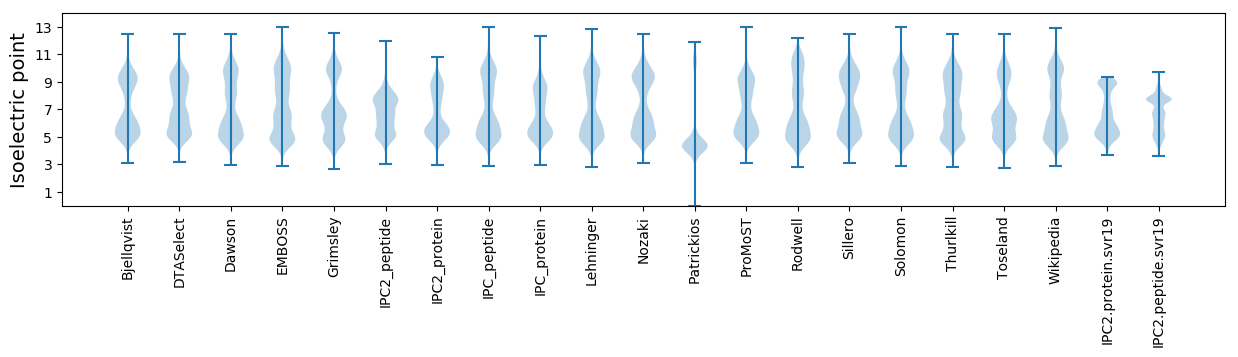
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8MV00|A0A5B8MV00_9CHLO Rad21/Rec8-like protein OS=Chloropicon primus OX=1764295 GN=A3770_12p66340 PE=3 SV=1
MM1 pKa = 6.93GHH3 pKa = 6.11KK4 pKa = 10.15NVWNSHH10 pKa = 4.99PRR12 pKa = 11.84DD13 pKa = 3.78YY14 pKa = 11.68GLGSHH19 pKa = 7.14PCRR22 pKa = 11.84VCGSHH27 pKa = 6.57HH28 pKa = 5.84GVIRR32 pKa = 11.84KK33 pKa = 9.4YY34 pKa = 10.89GLNICRR40 pKa = 11.84RR41 pKa = 11.84CFRR44 pKa = 11.84EE45 pKa = 3.58NAKK48 pKa = 10.78DD49 pKa = 2.91IGFRR53 pKa = 11.84KK54 pKa = 9.62LRR56 pKa = 3.83
MM1 pKa = 6.93GHH3 pKa = 6.11KK4 pKa = 10.15NVWNSHH10 pKa = 4.99PRR12 pKa = 11.84DD13 pKa = 3.78YY14 pKa = 11.68GLGSHH19 pKa = 7.14PCRR22 pKa = 11.84VCGSHH27 pKa = 6.57HH28 pKa = 5.84GVIRR32 pKa = 11.84KK33 pKa = 9.4YY34 pKa = 10.89GLNICRR40 pKa = 11.84RR41 pKa = 11.84CFRR44 pKa = 11.84EE45 pKa = 3.58NAKK48 pKa = 10.78DD49 pKa = 2.91IGFRR53 pKa = 11.84KK54 pKa = 9.62LRR56 pKa = 3.83
Molecular weight: 6.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4253573 |
46 |
11548 |
494.3 |
54.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.343 ± 0.028 | 1.757 ± 0.03 |
5.416 ± 0.018 | 7.653 ± 0.039 |
3.663 ± 0.016 | 7.856 ± 0.035 |
2.049 ± 0.013 | 3.987 ± 0.021 |
6.411 ± 0.033 | 9.297 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.012 | 3.389 ± 0.018 |
4.274 ± 0.02 | 3.796 ± 0.017 |
5.936 ± 0.03 | 7.929 ± 0.034 |
4.997 ± 0.023 | 7.038 ± 0.022 |
1.216 ± 0.01 | 2.569 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
