
Silurus glanis circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; European catfish circovirus
Average proteome isoelectric point is 8.88
Get precalculated fractions of proteins
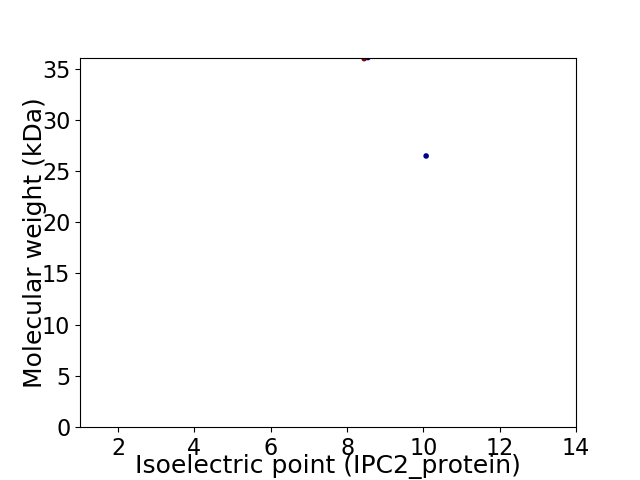
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1TEL3|I1TEL3_9CIRC ATP-dependent helicase Rep OS=Silurus glanis circovirus OX=1126411 GN=Rep PE=3 SV=1
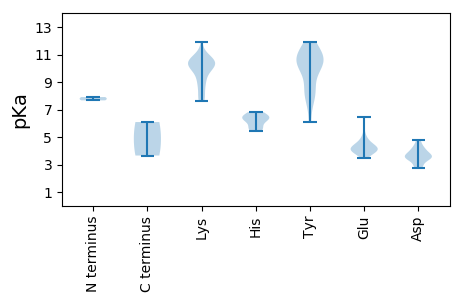
MM1 pKa = 7.91PKK3 pKa = 9.78YY4 pKa = 10.12QKK6 pKa = 10.49KK7 pKa = 10.37SPGKK11 pKa = 8.16EE12 pKa = 3.46NRR14 pKa = 11.84SPVQRR19 pKa = 11.84PKK21 pKa = 10.31QARR24 pKa = 11.84PPKK27 pKa = 10.2KK28 pKa = 8.74EE29 pKa = 4.09APHH32 pKa = 5.64KK33 pKa = 10.6RR34 pKa = 11.84YY35 pKa = 10.58VFTLNNYY42 pKa = 6.09TTEE45 pKa = 4.01EE46 pKa = 4.19YY47 pKa = 11.25ARR49 pKa = 11.84IDD51 pKa = 3.5NVGADD56 pKa = 3.37GLARR60 pKa = 11.84YY61 pKa = 7.49MITGKK66 pKa = 10.22EE67 pKa = 3.96VGEE70 pKa = 4.45NGTPHH75 pKa = 6.3LQGFINLKK83 pKa = 7.92VKK85 pKa = 10.44KK86 pKa = 9.73RR87 pKa = 11.84FSQIKK92 pKa = 8.89EE93 pKa = 3.9MLGSRR98 pKa = 11.84CHH100 pKa = 6.22IEE102 pKa = 3.89KK103 pKa = 10.8ARR105 pKa = 11.84GTDD108 pKa = 3.27LEE110 pKa = 4.07NRR112 pKa = 11.84VYY114 pKa = 10.49CSKK117 pKa = 10.7EE118 pKa = 3.69GSFQEE123 pKa = 4.31YY124 pKa = 10.05GSPVGQGKK132 pKa = 10.26RR133 pKa = 11.84SDD135 pKa = 3.55LDD137 pKa = 3.44AAAEE141 pKa = 4.11TLRR144 pKa = 11.84TSLGDD149 pKa = 3.5LRR151 pKa = 11.84SVAEE155 pKa = 4.72LYY157 pKa = 9.71PSQFIRR163 pKa = 11.84YY164 pKa = 8.32GRR166 pKa = 11.84GLRR169 pKa = 11.84DD170 pKa = 3.38YY171 pKa = 11.31ASVLGLVKK179 pKa = 10.53PRR181 pKa = 11.84DD182 pKa = 3.64FKK184 pKa = 11.93ANVTVITGPPGCGKK198 pKa = 10.03SRR200 pKa = 11.84YY201 pKa = 9.19AADD204 pKa = 3.77HH205 pKa = 6.63ASGTPYY211 pKa = 10.56YY212 pKa = 10.2KK213 pKa = 10.28PRR215 pKa = 11.84GDD217 pKa = 2.86WWDD220 pKa = 4.1GYY222 pKa = 8.5HH223 pKa = 6.5TNATVILDD231 pKa = 4.8DD232 pKa = 4.05FYY234 pKa = 11.92GWIKK238 pKa = 10.27LDD240 pKa = 3.44EE241 pKa = 4.24MLRR244 pKa = 11.84ICDD247 pKa = 4.21RR248 pKa = 11.84YY249 pKa = 8.57PHH251 pKa = 5.45QVPVKK256 pKa = 9.28GGYY259 pKa = 7.91VQFLARR265 pKa = 11.84DD266 pKa = 3.48IFITSNKK273 pKa = 8.76PVEE276 pKa = 4.2EE277 pKa = 4.19WFPNCDD283 pKa = 2.87CSALFRR289 pKa = 11.84RR290 pKa = 11.84INVYY294 pKa = 7.92LTWNDD299 pKa = 2.75EE300 pKa = 4.39CFVEE304 pKa = 6.43RR305 pKa = 11.84IDD307 pKa = 3.68TPYY310 pKa = 10.74EE311 pKa = 3.83INFF314 pKa = 3.65
MM1 pKa = 7.91PKK3 pKa = 9.78YY4 pKa = 10.12QKK6 pKa = 10.49KK7 pKa = 10.37SPGKK11 pKa = 8.16EE12 pKa = 3.46NRR14 pKa = 11.84SPVQRR19 pKa = 11.84PKK21 pKa = 10.31QARR24 pKa = 11.84PPKK27 pKa = 10.2KK28 pKa = 8.74EE29 pKa = 4.09APHH32 pKa = 5.64KK33 pKa = 10.6RR34 pKa = 11.84YY35 pKa = 10.58VFTLNNYY42 pKa = 6.09TTEE45 pKa = 4.01EE46 pKa = 4.19YY47 pKa = 11.25ARR49 pKa = 11.84IDD51 pKa = 3.5NVGADD56 pKa = 3.37GLARR60 pKa = 11.84YY61 pKa = 7.49MITGKK66 pKa = 10.22EE67 pKa = 3.96VGEE70 pKa = 4.45NGTPHH75 pKa = 6.3LQGFINLKK83 pKa = 7.92VKK85 pKa = 10.44KK86 pKa = 9.73RR87 pKa = 11.84FSQIKK92 pKa = 8.89EE93 pKa = 3.9MLGSRR98 pKa = 11.84CHH100 pKa = 6.22IEE102 pKa = 3.89KK103 pKa = 10.8ARR105 pKa = 11.84GTDD108 pKa = 3.27LEE110 pKa = 4.07NRR112 pKa = 11.84VYY114 pKa = 10.49CSKK117 pKa = 10.7EE118 pKa = 3.69GSFQEE123 pKa = 4.31YY124 pKa = 10.05GSPVGQGKK132 pKa = 10.26RR133 pKa = 11.84SDD135 pKa = 3.55LDD137 pKa = 3.44AAAEE141 pKa = 4.11TLRR144 pKa = 11.84TSLGDD149 pKa = 3.5LRR151 pKa = 11.84SVAEE155 pKa = 4.72LYY157 pKa = 9.71PSQFIRR163 pKa = 11.84YY164 pKa = 8.32GRR166 pKa = 11.84GLRR169 pKa = 11.84DD170 pKa = 3.38YY171 pKa = 11.31ASVLGLVKK179 pKa = 10.53PRR181 pKa = 11.84DD182 pKa = 3.64FKK184 pKa = 11.93ANVTVITGPPGCGKK198 pKa = 10.03SRR200 pKa = 11.84YY201 pKa = 9.19AADD204 pKa = 3.77HH205 pKa = 6.63ASGTPYY211 pKa = 10.56YY212 pKa = 10.2KK213 pKa = 10.28PRR215 pKa = 11.84GDD217 pKa = 2.86WWDD220 pKa = 4.1GYY222 pKa = 8.5HH223 pKa = 6.5TNATVILDD231 pKa = 4.8DD232 pKa = 4.05FYY234 pKa = 11.92GWIKK238 pKa = 10.27LDD240 pKa = 3.44EE241 pKa = 4.24MLRR244 pKa = 11.84ICDD247 pKa = 4.21RR248 pKa = 11.84YY249 pKa = 8.57PHH251 pKa = 5.45QVPVKK256 pKa = 9.28GGYY259 pKa = 7.91VQFLARR265 pKa = 11.84DD266 pKa = 3.48IFITSNKK273 pKa = 8.76PVEE276 pKa = 4.2EE277 pKa = 4.19WFPNCDD283 pKa = 2.87CSALFRR289 pKa = 11.84RR290 pKa = 11.84INVYY294 pKa = 7.92LTWNDD299 pKa = 2.75EE300 pKa = 4.39CFVEE304 pKa = 6.43RR305 pKa = 11.84IDD307 pKa = 3.68TPYY310 pKa = 10.74EE311 pKa = 3.83INFF314 pKa = 3.65
Molecular weight: 35.96 kDa
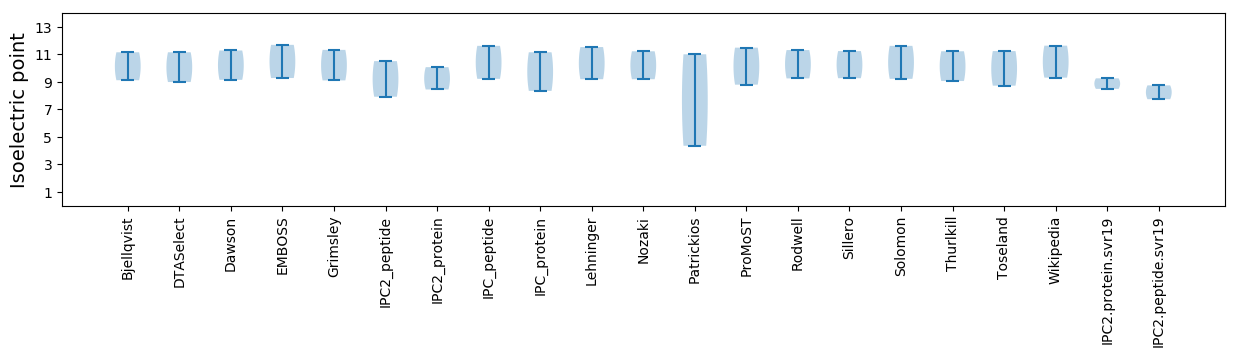
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1TEL3|I1TEL3_9CIRC ATP-dependent helicase Rep OS=Silurus glanis circovirus OX=1126411 GN=Rep PE=3 SV=1
MM1 pKa = 7.67AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TFRR9 pKa = 11.84RR10 pKa = 11.84PIRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84MHH17 pKa = 5.71RR18 pKa = 11.84RR19 pKa = 11.84TRR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84MIRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84SRR34 pKa = 11.84PMGMRR39 pKa = 11.84SFNFRR44 pKa = 11.84AIKK47 pKa = 10.13LDD49 pKa = 3.9HH50 pKa = 6.81IKK52 pKa = 10.94VGGNSSVTGVLTFCLKK68 pKa = 10.65DD69 pKa = 3.69FCANFLNWDD78 pKa = 3.7YY79 pKa = 11.72YY80 pKa = 10.72RR81 pKa = 11.84FNAVVVKK88 pKa = 7.75FVPQNTAQFDD98 pKa = 3.9QEE100 pKa = 4.62GGLSTVCATAVDD112 pKa = 4.19YY113 pKa = 11.29DD114 pKa = 4.63DD115 pKa = 4.37EE116 pKa = 4.79SKK118 pKa = 9.26PTSVRR123 pKa = 11.84QLMMMQNARR132 pKa = 11.84TFRR135 pKa = 11.84TGVGHH140 pKa = 6.28SRR142 pKa = 11.84KK143 pKa = 9.12FKK145 pKa = 10.26PKK147 pKa = 7.63MTFSVVEE154 pKa = 4.22KK155 pKa = 10.01VDD157 pKa = 3.63SVTTYY162 pKa = 11.33APAGLWMGGNRR173 pKa = 11.84KK174 pKa = 7.67WWLNTGYY181 pKa = 10.53SEE183 pKa = 5.18VKK185 pKa = 10.51FLGLKK190 pKa = 10.17YY191 pKa = 10.9AMQNMSANDD200 pKa = 2.86IHH202 pKa = 6.46YY203 pKa = 10.1QVLIKK208 pKa = 10.71GYY210 pKa = 10.39FSFKK214 pKa = 10.68SPLITAPTVRR224 pKa = 11.84GEE226 pKa = 4.04HH227 pKa = 6.09
MM1 pKa = 7.67AFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84TFRR9 pKa = 11.84RR10 pKa = 11.84PIRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84MHH17 pKa = 5.71RR18 pKa = 11.84RR19 pKa = 11.84TRR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84MIRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84SRR34 pKa = 11.84PMGMRR39 pKa = 11.84SFNFRR44 pKa = 11.84AIKK47 pKa = 10.13LDD49 pKa = 3.9HH50 pKa = 6.81IKK52 pKa = 10.94VGGNSSVTGVLTFCLKK68 pKa = 10.65DD69 pKa = 3.69FCANFLNWDD78 pKa = 3.7YY79 pKa = 11.72YY80 pKa = 10.72RR81 pKa = 11.84FNAVVVKK88 pKa = 7.75FVPQNTAQFDD98 pKa = 3.9QEE100 pKa = 4.62GGLSTVCATAVDD112 pKa = 4.19YY113 pKa = 11.29DD114 pKa = 4.63DD115 pKa = 4.37EE116 pKa = 4.79SKK118 pKa = 9.26PTSVRR123 pKa = 11.84QLMMMQNARR132 pKa = 11.84TFRR135 pKa = 11.84TGVGHH140 pKa = 6.28SRR142 pKa = 11.84KK143 pKa = 9.12FKK145 pKa = 10.26PKK147 pKa = 7.63MTFSVVEE154 pKa = 4.22KK155 pKa = 10.01VDD157 pKa = 3.63SVTTYY162 pKa = 11.33APAGLWMGGNRR173 pKa = 11.84KK174 pKa = 7.67WWLNTGYY181 pKa = 10.53SEE183 pKa = 5.18VKK185 pKa = 10.51FLGLKK190 pKa = 10.17YY191 pKa = 10.9AMQNMSANDD200 pKa = 2.86IHH202 pKa = 6.46YY203 pKa = 10.1QVLIKK208 pKa = 10.71GYY210 pKa = 10.39FSFKK214 pKa = 10.68SPLITAPTVRR224 pKa = 11.84GEE226 pKa = 4.04HH227 pKa = 6.09
Molecular weight: 26.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
541 |
227 |
314 |
270.5 |
31.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.002 | 1.848 ± 0.344 |
5.176 ± 0.791 | 4.436 ± 1.459 |
5.36 ± 1.103 | 7.763 ± 0.467 |
2.033 ± 0.111 | 4.067 ± 0.642 |
6.839 ± 0.439 | 5.915 ± 0.411 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.957 ± 1.521 | 4.621 ± 0.147 |
5.176 ± 1.079 | 3.142 ± 0.038 |
9.797 ± 1.658 | 5.73 ± 0.573 |
5.915 ± 0.74 | 6.839 ± 0.712 |
1.664 ± 0.064 | 4.991 ± 0.958 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
