
Elderberry carlavirus A
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Carlavirus; unclassified Carlavirus
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
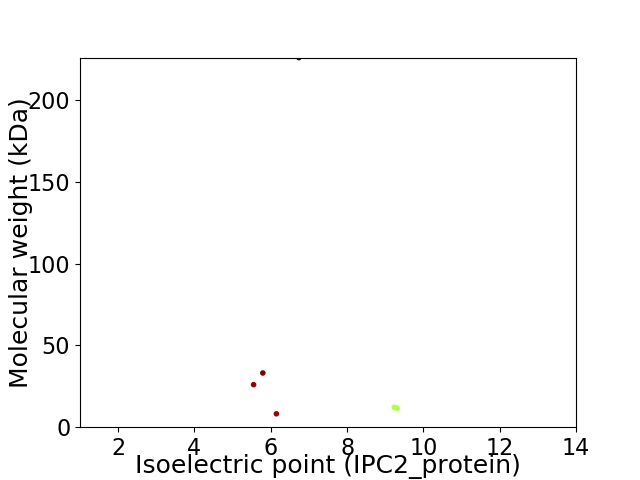
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
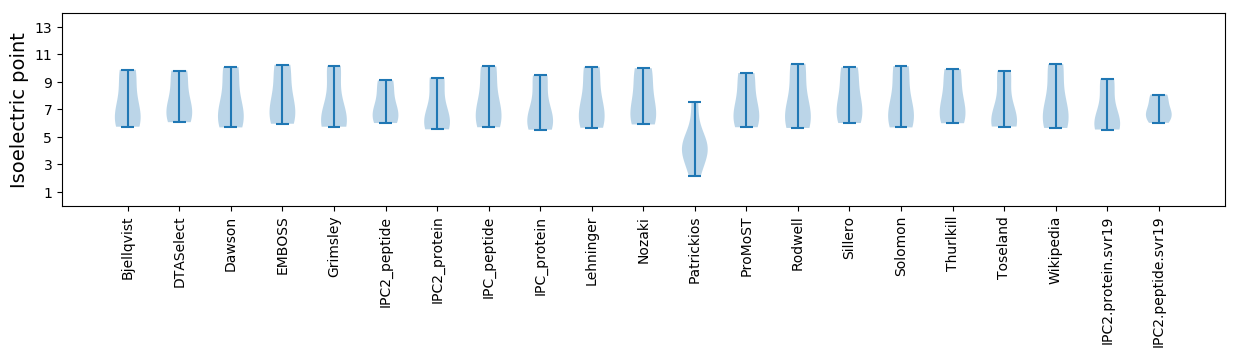
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7MDS4|A0A0A7MDS4_9VIRU Triple gene block protein 1 OS=Elderberry carlavirus A OX=1569052 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 4.21VLIKK6 pKa = 10.59LALEE10 pKa = 4.1RR11 pKa = 11.84GFDD14 pKa = 4.26RR15 pKa = 11.84IHH17 pKa = 6.89SNYY20 pKa = 9.77SGCAVFHH27 pKa = 6.19CVPGAGKK34 pKa = 6.92TTLIRR39 pKa = 11.84DD40 pKa = 4.13LLQRR44 pKa = 11.84DD45 pKa = 3.68SRR47 pKa = 11.84FQALTFGVVDD57 pKa = 3.86KK58 pKa = 11.21QSLSGRR64 pKa = 11.84RR65 pKa = 11.84IKK67 pKa = 10.9GPSEE71 pKa = 3.57RR72 pKa = 11.84LEE74 pKa = 4.11EE75 pKa = 4.39GKK77 pKa = 8.59LTIVDD82 pKa = 4.71EE83 pKa = 4.33YY84 pKa = 11.52TEE86 pKa = 4.65GDD88 pKa = 3.23WEE90 pKa = 4.36IYY92 pKa = 10.37KK93 pKa = 10.35PIALFGDD100 pKa = 5.22PIQSCNLSRR109 pKa = 11.84VKK111 pKa = 10.39PSNFEE116 pKa = 3.67CRR118 pKa = 11.84RR119 pKa = 11.84TFRR122 pKa = 11.84FGRR125 pKa = 11.84QTCALLGRR133 pKa = 11.84LGFHH137 pKa = 6.41ITSEE141 pKa = 4.12FDD143 pKa = 3.49DD144 pKa = 4.04EE145 pKa = 5.32VIIGDD150 pKa = 3.76VFSIEE155 pKa = 4.06PEE157 pKa = 3.92GLIIAFEE164 pKa = 4.35PEE166 pKa = 4.19VKK168 pKa = 10.28EE169 pKa = 3.76LLEE172 pKa = 4.62DD173 pKa = 3.76HH174 pKa = 6.63GCDD177 pKa = 3.58YY178 pKa = 11.48SEE180 pKa = 4.48PCALRR185 pKa = 11.84GRR187 pKa = 11.84TTKK190 pKa = 10.73AVTFITASEE199 pKa = 4.62SIPEE203 pKa = 4.14SLAHH207 pKa = 6.62LFFIALTRR215 pKa = 11.84HH216 pKa = 4.23QEE218 pKa = 3.89KK219 pKa = 10.84LIVLSRR225 pKa = 11.84NATFGATT232 pKa = 3.31
MM1 pKa = 7.98DD2 pKa = 4.21VLIKK6 pKa = 10.59LALEE10 pKa = 4.1RR11 pKa = 11.84GFDD14 pKa = 4.26RR15 pKa = 11.84IHH17 pKa = 6.89SNYY20 pKa = 9.77SGCAVFHH27 pKa = 6.19CVPGAGKK34 pKa = 6.92TTLIRR39 pKa = 11.84DD40 pKa = 4.13LLQRR44 pKa = 11.84DD45 pKa = 3.68SRR47 pKa = 11.84FQALTFGVVDD57 pKa = 3.86KK58 pKa = 11.21QSLSGRR64 pKa = 11.84RR65 pKa = 11.84IKK67 pKa = 10.9GPSEE71 pKa = 3.57RR72 pKa = 11.84LEE74 pKa = 4.11EE75 pKa = 4.39GKK77 pKa = 8.59LTIVDD82 pKa = 4.71EE83 pKa = 4.33YY84 pKa = 11.52TEE86 pKa = 4.65GDD88 pKa = 3.23WEE90 pKa = 4.36IYY92 pKa = 10.37KK93 pKa = 10.35PIALFGDD100 pKa = 5.22PIQSCNLSRR109 pKa = 11.84VKK111 pKa = 10.39PSNFEE116 pKa = 3.67CRR118 pKa = 11.84RR119 pKa = 11.84TFRR122 pKa = 11.84FGRR125 pKa = 11.84QTCALLGRR133 pKa = 11.84LGFHH137 pKa = 6.41ITSEE141 pKa = 4.12FDD143 pKa = 3.49DD144 pKa = 4.04EE145 pKa = 5.32VIIGDD150 pKa = 3.76VFSIEE155 pKa = 4.06PEE157 pKa = 3.92GLIIAFEE164 pKa = 4.35PEE166 pKa = 4.19VKK168 pKa = 10.28EE169 pKa = 3.76LLEE172 pKa = 4.62DD173 pKa = 3.76HH174 pKa = 6.63GCDD177 pKa = 3.58YY178 pKa = 11.48SEE180 pKa = 4.48PCALRR185 pKa = 11.84GRR187 pKa = 11.84TTKK190 pKa = 10.73AVTFITASEE199 pKa = 4.62SIPEE203 pKa = 4.14SLAHH207 pKa = 6.62LFFIALTRR215 pKa = 11.84HH216 pKa = 4.23QEE218 pKa = 3.89KK219 pKa = 10.84LIVLSRR225 pKa = 11.84NATFGATT232 pKa = 3.31
Molecular weight: 26.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7M910|A0A0A7M910_9VIRU Capsid protein OS=Elderberry carlavirus A OX=1569052 PE=3 SV=1
MM1 pKa = 7.84DD2 pKa = 4.79RR3 pKa = 11.84VKK5 pKa = 10.91LMCDD9 pKa = 3.71IIRR12 pKa = 11.84ILSSEE17 pKa = 4.01SRR19 pKa = 11.84EE20 pKa = 3.92AYY22 pKa = 10.41YY23 pKa = 10.13IDD25 pKa = 2.97IARR28 pKa = 11.84YY29 pKa = 8.59IVHH32 pKa = 7.12LSAEE36 pKa = 4.03LHH38 pKa = 6.45APGKK42 pKa = 9.11STFAKK47 pKa = 9.85RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AKK52 pKa = 10.69SLGRR56 pKa = 11.84CHH58 pKa = 6.31RR59 pKa = 11.84CYY61 pKa = 10.46RR62 pKa = 11.84ISPGFYY68 pKa = 8.89HH69 pKa = 5.52TTKK72 pKa = 10.56CDD74 pKa = 4.11GISCTDD80 pKa = 3.2SFGSRR85 pKa = 11.84SKK87 pKa = 9.68HH88 pKa = 5.23RR89 pKa = 11.84NYY91 pKa = 9.76IVHH94 pKa = 6.51GQRR97 pKa = 11.84KK98 pKa = 7.88HH99 pKa = 5.07CC100 pKa = 4.36
MM1 pKa = 7.84DD2 pKa = 4.79RR3 pKa = 11.84VKK5 pKa = 10.91LMCDD9 pKa = 3.71IIRR12 pKa = 11.84ILSSEE17 pKa = 4.01SRR19 pKa = 11.84EE20 pKa = 3.92AYY22 pKa = 10.41YY23 pKa = 10.13IDD25 pKa = 2.97IARR28 pKa = 11.84YY29 pKa = 8.59IVHH32 pKa = 7.12LSAEE36 pKa = 4.03LHH38 pKa = 6.45APGKK42 pKa = 9.11STFAKK47 pKa = 9.85RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AKK52 pKa = 10.69SLGRR56 pKa = 11.84CHH58 pKa = 6.31RR59 pKa = 11.84CYY61 pKa = 10.46RR62 pKa = 11.84ISPGFYY68 pKa = 8.89HH69 pKa = 5.52TTKK72 pKa = 10.56CDD74 pKa = 4.11GISCTDD80 pKa = 3.2SFGSRR85 pKa = 11.84SKK87 pKa = 9.68HH88 pKa = 5.23RR89 pKa = 11.84NYY91 pKa = 9.76IVHH94 pKa = 6.51GQRR97 pKa = 11.84KK98 pKa = 7.88HH99 pKa = 5.07CC100 pKa = 4.36
Molecular weight: 11.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2807 |
72 |
1995 |
467.8 |
52.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.27 ± 0.587 | 3.206 ± 0.389 |
4.881 ± 0.345 | 6.947 ± 1.084 |
5.522 ± 0.854 | 6.056 ± 0.507 |
2.458 ± 0.706 | 5.949 ± 0.555 |
6.484 ± 0.599 | 10.046 ± 1.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.031 ± 0.336 | 4.239 ± 0.558 |
4.275 ± 0.628 | 2.814 ± 0.462 |
5.878 ± 0.985 | 7.731 ± 0.685 |
5.166 ± 0.555 | 5.771 ± 0.497 |
0.891 ± 0.184 | 3.384 ± 0.662 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
