
Cellulophaga phage phi18:3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
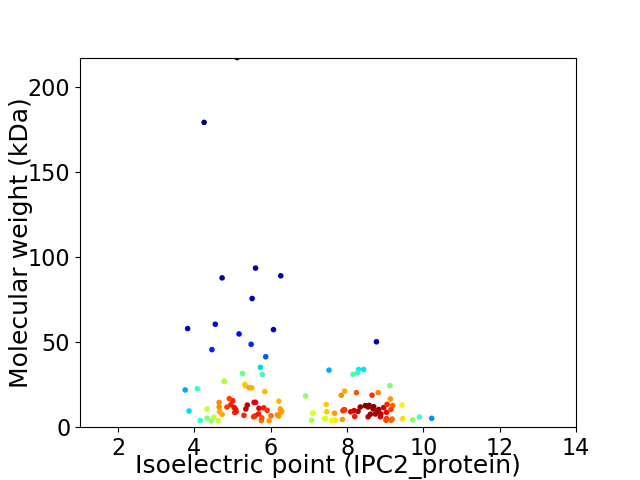
Virtual 2D-PAGE plot for 123 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
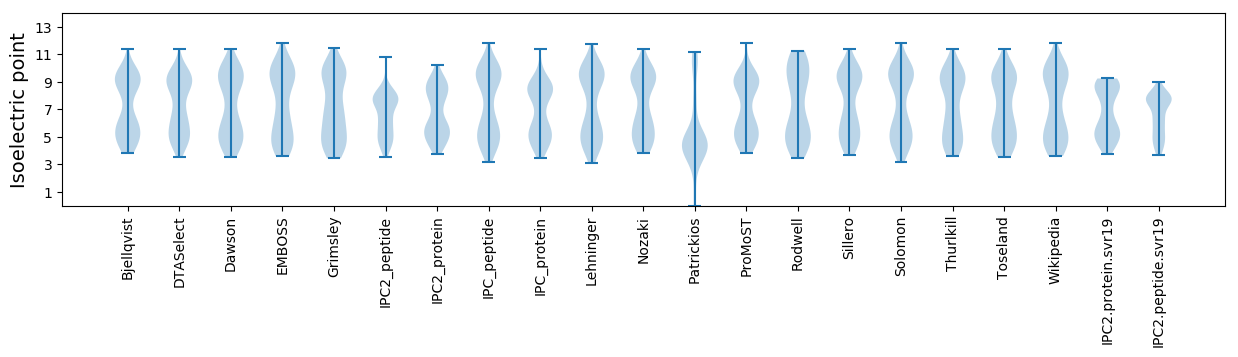
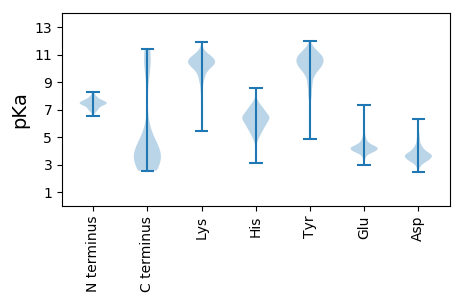
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0A2B7|S0A2B7_9CAUD Uncharacterized protein OS=Cellulophaga phage phi18:3 OX=1327983 GN=Phi18:3_gp095 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.61NKK4 pKa = 8.56ITIIMILMSISFGYY18 pKa = 9.85SQVLVPEE25 pKa = 4.2NTGYY29 pKa = 11.18GYY31 pKa = 11.07DD32 pKa = 3.53NLTTVQLNALTKK44 pKa = 10.45KK45 pKa = 10.43KK46 pKa = 10.94VGDD49 pKa = 3.23IYY51 pKa = 11.59YY52 pKa = 10.48NADD55 pKa = 3.03LGYY58 pKa = 9.03HH59 pKa = 5.2VKK61 pKa = 9.49WNGVIFEE68 pKa = 4.6SLASGISGIDD78 pKa = 3.43PADD81 pKa = 3.37QDD83 pKa = 3.85KK84 pKa = 11.33LNNISITQPVNLDD97 pKa = 3.69TVEE100 pKa = 4.31SLANSALQTEE110 pKa = 4.96TDD112 pKa = 3.69PTTTPANIKK121 pKa = 10.45AKK123 pKa = 10.55LEE125 pKa = 4.19SLTGTNRR132 pKa = 11.84LDD134 pKa = 3.27AFAIKK139 pKa = 10.27NLSAQEE145 pKa = 3.96TDD147 pKa = 3.46PNLTKK152 pKa = 11.0SNVEE156 pKa = 3.97GLGISYY162 pKa = 9.47TSLSDD167 pKa = 3.67VPSVSPPLTFGPGFTRR183 pKa = 11.84TGDD186 pKa = 3.71NIVLDD191 pKa = 4.09NPFTTANEE199 pKa = 4.21TTLNSALQSEE209 pKa = 4.65TDD211 pKa = 3.7PLYY214 pKa = 10.96SAWDD218 pKa = 3.41KK219 pKa = 11.41DD220 pKa = 4.07YY221 pKa = 11.58NDD223 pKa = 5.36LINRR227 pKa = 11.84PVNLVTYY234 pKa = 10.27DD235 pKa = 3.75PLTAQDD241 pKa = 3.55VQISVLGGSDD251 pKa = 3.46SEE253 pKa = 4.81SYY255 pKa = 10.68ILEE258 pKa = 4.39FDD260 pKa = 3.74PDD262 pKa = 3.92GYY264 pKa = 10.94DD265 pKa = 3.42YY266 pKa = 11.44SGEE269 pKa = 3.95PSGTGTVSQTIIDD282 pKa = 4.05GDD284 pKa = 4.09TNAVSGNAVFDD295 pKa = 3.89GLATKK300 pKa = 10.28LQAGSNDD307 pKa = 3.43LGSSNFYY314 pKa = 10.92VNSDD318 pKa = 4.2GILALANSNTQSTVFVNPYY337 pKa = 10.2GSGLIAGNSDD347 pKa = 3.19DD348 pKa = 4.6SEE350 pKa = 4.28IMKK353 pKa = 9.18LTLSPSSITADD364 pKa = 3.66LSNSQITSTGATSLITKK381 pKa = 9.4GYY383 pKa = 10.9ADD385 pKa = 3.54ATYY388 pKa = 10.93ASSNQNLSLDD398 pKa = 3.5MGYY401 pKa = 11.01GILSISGGNSVDD413 pKa = 4.2LSATSSYY420 pKa = 11.09TSFDD424 pKa = 3.19IVDD427 pKa = 3.77TGSGATYY434 pKa = 10.36AKK436 pKa = 10.41SGVQEE441 pKa = 3.92KK442 pKa = 11.07DD443 pKa = 3.43LITEE447 pKa = 4.54LSQNNIILSSEE458 pKa = 4.23TNTVSGTPSGNLRR471 pKa = 11.84ILLQTYY477 pKa = 7.76YY478 pKa = 10.95VQTEE482 pKa = 4.44FKK484 pKa = 10.26TFFDD488 pKa = 4.03VFIEE492 pKa = 4.0NMNGVGDD499 pKa = 4.14YY500 pKa = 10.0YY501 pKa = 11.41LKK503 pKa = 11.08GILNPAGTQIDD514 pKa = 4.71VYY516 pKa = 11.19SVGSDD521 pKa = 3.21GLLTPVSAPTSLGYY535 pKa = 9.93IQITGTYY542 pKa = 8.97FYY544 pKa = 11.31AGFF547 pKa = 4.16
MM1 pKa = 7.47KK2 pKa = 10.61NKK4 pKa = 8.56ITIIMILMSISFGYY18 pKa = 9.85SQVLVPEE25 pKa = 4.2NTGYY29 pKa = 11.18GYY31 pKa = 11.07DD32 pKa = 3.53NLTTVQLNALTKK44 pKa = 10.45KK45 pKa = 10.43KK46 pKa = 10.94VGDD49 pKa = 3.23IYY51 pKa = 11.59YY52 pKa = 10.48NADD55 pKa = 3.03LGYY58 pKa = 9.03HH59 pKa = 5.2VKK61 pKa = 9.49WNGVIFEE68 pKa = 4.6SLASGISGIDD78 pKa = 3.43PADD81 pKa = 3.37QDD83 pKa = 3.85KK84 pKa = 11.33LNNISITQPVNLDD97 pKa = 3.69TVEE100 pKa = 4.31SLANSALQTEE110 pKa = 4.96TDD112 pKa = 3.69PTTTPANIKK121 pKa = 10.45AKK123 pKa = 10.55LEE125 pKa = 4.19SLTGTNRR132 pKa = 11.84LDD134 pKa = 3.27AFAIKK139 pKa = 10.27NLSAQEE145 pKa = 3.96TDD147 pKa = 3.46PNLTKK152 pKa = 11.0SNVEE156 pKa = 3.97GLGISYY162 pKa = 9.47TSLSDD167 pKa = 3.67VPSVSPPLTFGPGFTRR183 pKa = 11.84TGDD186 pKa = 3.71NIVLDD191 pKa = 4.09NPFTTANEE199 pKa = 4.21TTLNSALQSEE209 pKa = 4.65TDD211 pKa = 3.7PLYY214 pKa = 10.96SAWDD218 pKa = 3.41KK219 pKa = 11.41DD220 pKa = 4.07YY221 pKa = 11.58NDD223 pKa = 5.36LINRR227 pKa = 11.84PVNLVTYY234 pKa = 10.27DD235 pKa = 3.75PLTAQDD241 pKa = 3.55VQISVLGGSDD251 pKa = 3.46SEE253 pKa = 4.81SYY255 pKa = 10.68ILEE258 pKa = 4.39FDD260 pKa = 3.74PDD262 pKa = 3.92GYY264 pKa = 10.94DD265 pKa = 3.42YY266 pKa = 11.44SGEE269 pKa = 3.95PSGTGTVSQTIIDD282 pKa = 4.05GDD284 pKa = 4.09TNAVSGNAVFDD295 pKa = 3.89GLATKK300 pKa = 10.28LQAGSNDD307 pKa = 3.43LGSSNFYY314 pKa = 10.92VNSDD318 pKa = 4.2GILALANSNTQSTVFVNPYY337 pKa = 10.2GSGLIAGNSDD347 pKa = 3.19DD348 pKa = 4.6SEE350 pKa = 4.28IMKK353 pKa = 9.18LTLSPSSITADD364 pKa = 3.66LSNSQITSTGATSLITKK381 pKa = 9.4GYY383 pKa = 10.9ADD385 pKa = 3.54ATYY388 pKa = 10.93ASSNQNLSLDD398 pKa = 3.5MGYY401 pKa = 11.01GILSISGGNSVDD413 pKa = 4.2LSATSSYY420 pKa = 11.09TSFDD424 pKa = 3.19IVDD427 pKa = 3.77TGSGATYY434 pKa = 10.36AKK436 pKa = 10.41SGVQEE441 pKa = 3.92KK442 pKa = 11.07DD443 pKa = 3.43LITEE447 pKa = 4.54LSQNNIILSSEE458 pKa = 4.23TNTVSGTPSGNLRR471 pKa = 11.84ILLQTYY477 pKa = 7.76YY478 pKa = 10.95VQTEE482 pKa = 4.44FKK484 pKa = 10.26TFFDD488 pKa = 4.03VFIEE492 pKa = 4.0NMNGVGDD499 pKa = 4.14YY500 pKa = 10.0YY501 pKa = 11.41LKK503 pKa = 11.08GILNPAGTQIDD514 pKa = 4.71VYY516 pKa = 11.19SVGSDD521 pKa = 3.21GLLTPVSAPTSLGYY535 pKa = 9.93IQITGTYY542 pKa = 8.97FYY544 pKa = 11.31AGFF547 pKa = 4.16
Molecular weight: 58.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9ZZ49|R9ZZ49_9CAUD Uncharacterized protein OS=Cellulophaga phage phi18:3 OX=1327983 GN=Phi18:3_gp107 PE=4 SV=1
MM1 pKa = 6.87EE2 pKa = 4.61QKK4 pKa = 10.63NPSRR8 pKa = 11.84LGRR11 pKa = 11.84VLNITGRR18 pKa = 11.84YY19 pKa = 8.75IEE21 pKa = 4.72LKK23 pKa = 10.36KK24 pKa = 11.02ASTKK28 pKa = 11.02DD29 pKa = 3.54NINIRR34 pKa = 11.84YY35 pKa = 9.18ISS37 pKa = 3.34
MM1 pKa = 6.87EE2 pKa = 4.61QKK4 pKa = 10.63NPSRR8 pKa = 11.84LGRR11 pKa = 11.84VLNITGRR18 pKa = 11.84YY19 pKa = 8.75IEE21 pKa = 4.72LKK23 pKa = 10.36KK24 pKa = 11.02ASTKK28 pKa = 11.02DD29 pKa = 3.54NINIRR34 pKa = 11.84YY35 pKa = 9.18ISS37 pKa = 3.34
Molecular weight: 4.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
22253 |
33 |
1935 |
180.9 |
20.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.087 ± 0.248 | 0.966 ± 0.124 |
6.453 ± 0.193 | 7.891 ± 0.302 |
4.624 ± 0.199 | 5.81 ± 0.285 |
1.294 ± 0.167 | 7.878 ± 0.218 |
9.338 ± 0.462 | 8.089 ± 0.231 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.292 ± 0.166 | 6.817 ± 0.311 |
2.741 ± 0.136 | 3.114 ± 0.206 |
3.685 ± 0.199 | 6.925 ± 0.236 |
6.228 ± 0.312 | 5.635 ± 0.169 |
1.025 ± 0.097 | 4.107 ± 0.15 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
