
Bryocella elongata
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Bryocella
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
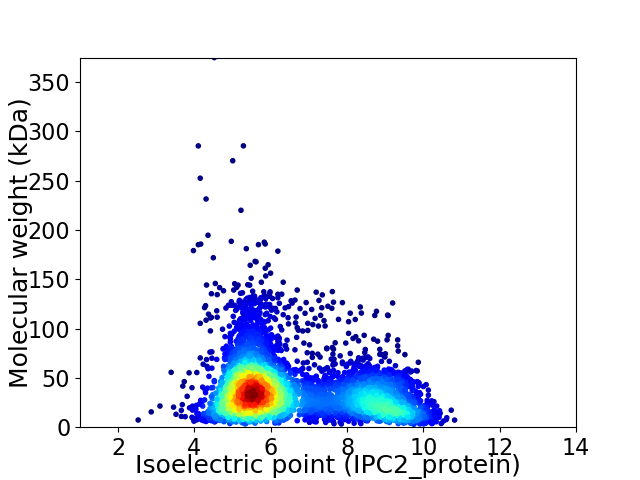
Virtual 2D-PAGE plot for 4503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
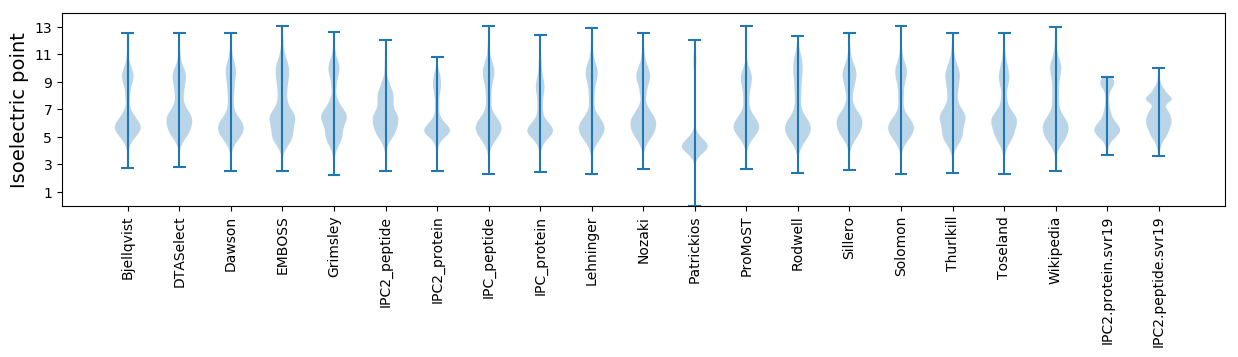
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H5ZFJ5|A0A1H5ZFJ5_9BACT Nucleobase:cation symporter-1 NCS1 family OS=Bryocella elongata OX=863522 GN=SAMN05421819_2636 PE=3 SV=1
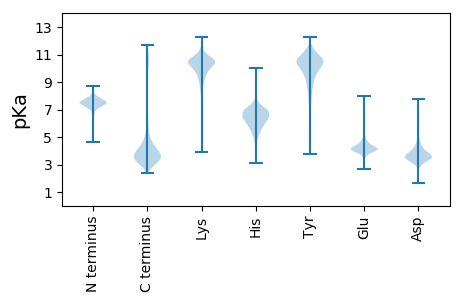
MM1 pKa = 7.45TPQEE5 pKa = 3.92QQLIDD10 pKa = 3.71GLISRR15 pKa = 11.84IRR17 pKa = 11.84STPATDD23 pKa = 3.88KK24 pKa = 11.44DD25 pKa = 3.55VDD27 pKa = 3.54ADD29 pKa = 4.25KK30 pKa = 11.25YY31 pKa = 10.94LHH33 pKa = 6.34QNLDD37 pKa = 3.58SVPDD41 pKa = 3.62VLYY44 pKa = 11.26VLAQTVLVQQYY55 pKa = 10.07GLQNAQQQLQAAQQQADD72 pKa = 4.04DD73 pKa = 4.68LQAQLDD79 pKa = 3.79QARR82 pKa = 11.84AQAQQAQQSQKK93 pKa = 9.64SGSFLGHH100 pKa = 7.46LFGTDD105 pKa = 3.01NPQPQQGYY113 pKa = 7.61PQQGYY118 pKa = 7.65PQGYY122 pKa = 7.82PPQGYY127 pKa = 8.65AQPGYY132 pKa = 8.6VQAVPVQPMGYY143 pKa = 10.67GMGGGMGSPIGGGGFLRR160 pKa = 11.84GALQTAAGVAAGEE173 pKa = 4.33MVFQGMEE180 pKa = 3.72DD181 pKa = 3.3MFRR184 pKa = 11.84GFGHH188 pKa = 6.77EE189 pKa = 4.01GGGYY193 pKa = 9.42GGSRR197 pKa = 11.84PTEE200 pKa = 3.97VINNNYY206 pKa = 9.75YY207 pKa = 10.91DD208 pKa = 4.19DD209 pKa = 5.0DD210 pKa = 4.84RR211 pKa = 11.84PEE213 pKa = 4.47SGHH216 pKa = 5.17EE217 pKa = 3.77HH218 pKa = 6.24HH219 pKa = 7.44LEE221 pKa = 4.3GNDD224 pKa = 3.95DD225 pKa = 3.73SSFYY229 pKa = 11.02NAANDD234 pKa = 3.91ASSKK238 pKa = 10.75DD239 pKa = 3.77LSPDD243 pKa = 2.96IEE245 pKa = 4.32DD246 pKa = 3.4RR247 pKa = 11.84RR248 pKa = 11.84GFADD252 pKa = 3.83TGNNDD257 pKa = 4.07ANDD260 pKa = 4.21DD261 pKa = 3.99SQQGFDD267 pKa = 4.91DD268 pKa = 4.54NSSSDD273 pKa = 3.73DD274 pKa = 3.59SGFDD278 pKa = 4.58DD279 pKa = 5.16SSSDD283 pKa = 4.29DD284 pKa = 3.36GGGYY288 pKa = 10.26DD289 pKa = 5.06DD290 pKa = 5.39SSNN293 pKa = 3.22
MM1 pKa = 7.45TPQEE5 pKa = 3.92QQLIDD10 pKa = 3.71GLISRR15 pKa = 11.84IRR17 pKa = 11.84STPATDD23 pKa = 3.88KK24 pKa = 11.44DD25 pKa = 3.55VDD27 pKa = 3.54ADD29 pKa = 4.25KK30 pKa = 11.25YY31 pKa = 10.94LHH33 pKa = 6.34QNLDD37 pKa = 3.58SVPDD41 pKa = 3.62VLYY44 pKa = 11.26VLAQTVLVQQYY55 pKa = 10.07GLQNAQQQLQAAQQQADD72 pKa = 4.04DD73 pKa = 4.68LQAQLDD79 pKa = 3.79QARR82 pKa = 11.84AQAQQAQQSQKK93 pKa = 9.64SGSFLGHH100 pKa = 7.46LFGTDD105 pKa = 3.01NPQPQQGYY113 pKa = 7.61PQQGYY118 pKa = 7.65PQGYY122 pKa = 7.82PPQGYY127 pKa = 8.65AQPGYY132 pKa = 8.6VQAVPVQPMGYY143 pKa = 10.67GMGGGMGSPIGGGGFLRR160 pKa = 11.84GALQTAAGVAAGEE173 pKa = 4.33MVFQGMEE180 pKa = 3.72DD181 pKa = 3.3MFRR184 pKa = 11.84GFGHH188 pKa = 6.77EE189 pKa = 4.01GGGYY193 pKa = 9.42GGSRR197 pKa = 11.84PTEE200 pKa = 3.97VINNNYY206 pKa = 9.75YY207 pKa = 10.91DD208 pKa = 4.19DD209 pKa = 5.0DD210 pKa = 4.84RR211 pKa = 11.84PEE213 pKa = 4.47SGHH216 pKa = 5.17EE217 pKa = 3.77HH218 pKa = 6.24HH219 pKa = 7.44LEE221 pKa = 4.3GNDD224 pKa = 3.95DD225 pKa = 3.73SSFYY229 pKa = 11.02NAANDD234 pKa = 3.91ASSKK238 pKa = 10.75DD239 pKa = 3.77LSPDD243 pKa = 2.96IEE245 pKa = 4.32DD246 pKa = 3.4RR247 pKa = 11.84RR248 pKa = 11.84GFADD252 pKa = 3.83TGNNDD257 pKa = 4.07ANDD260 pKa = 4.21DD261 pKa = 3.99SQQGFDD267 pKa = 4.91DD268 pKa = 4.54NSSSDD273 pKa = 3.73DD274 pKa = 3.59SGFDD278 pKa = 4.58DD279 pKa = 5.16SSSDD283 pKa = 4.29DD284 pKa = 3.36GGGYY288 pKa = 10.26DD289 pKa = 5.06DD290 pKa = 5.39SSNN293 pKa = 3.22
Molecular weight: 31.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H5WUH4|A0A1H5WUH4_9BACT Uncharacterized protein OS=Bryocella elongata OX=863522 GN=SAMN05421819_1730 PE=4 SV=1
MM1 pKa = 6.72TRR3 pKa = 11.84KK4 pKa = 9.59SIKK7 pKa = 8.64IHH9 pKa = 6.27KK10 pKa = 7.38PAPTLHH16 pKa = 6.66LPAHH20 pKa = 5.77EE21 pKa = 4.01QRR23 pKa = 11.84AAARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84HH30 pKa = 5.6NAALPNIPGPKK41 pKa = 9.02PRR43 pKa = 11.84IARR46 pKa = 11.84AAKK49 pKa = 8.89PRR51 pKa = 11.84PIPAHH56 pKa = 6.44KK57 pKa = 9.4VTTPRR62 pKa = 11.84SVHH65 pKa = 5.53
MM1 pKa = 6.72TRR3 pKa = 11.84KK4 pKa = 9.59SIKK7 pKa = 8.64IHH9 pKa = 6.27KK10 pKa = 7.38PAPTLHH16 pKa = 6.66LPAHH20 pKa = 5.77EE21 pKa = 4.01QRR23 pKa = 11.84AAARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84HH30 pKa = 5.6NAALPNIPGPKK41 pKa = 9.02PRR43 pKa = 11.84IARR46 pKa = 11.84AAKK49 pKa = 8.89PRR51 pKa = 11.84PIPAHH56 pKa = 6.44KK57 pKa = 9.4VTTPRR62 pKa = 11.84SVHH65 pKa = 5.53
Molecular weight: 7.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1673813 |
25 |
3776 |
371.7 |
40.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.471 ± 0.051 | 0.857 ± 0.011 |
5.116 ± 0.031 | 5.317 ± 0.048 |
3.788 ± 0.02 | 8.146 ± 0.046 |
2.327 ± 0.017 | 4.703 ± 0.027 |
3.417 ± 0.03 | 9.895 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.301 ± 0.018 | 3.259 ± 0.033 |
5.477 ± 0.029 | 3.677 ± 0.026 |
6.072 ± 0.04 | 6.464 ± 0.042 |
6.346 ± 0.056 | 7.246 ± 0.03 |
1.384 ± 0.018 | 2.738 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
