
Gyrovirus Tu243
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Gyrovirus homsa4
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
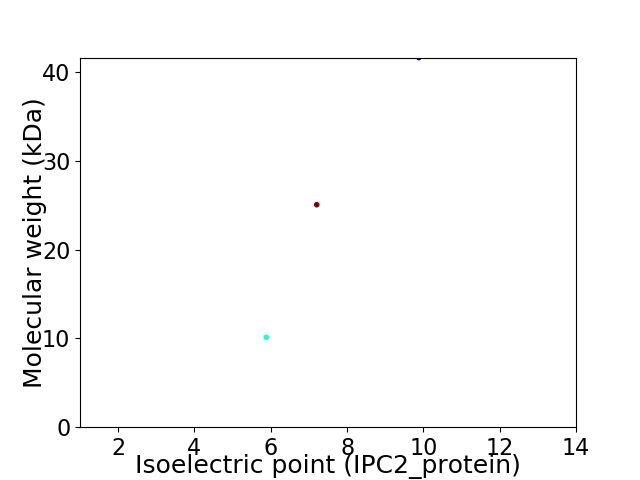
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
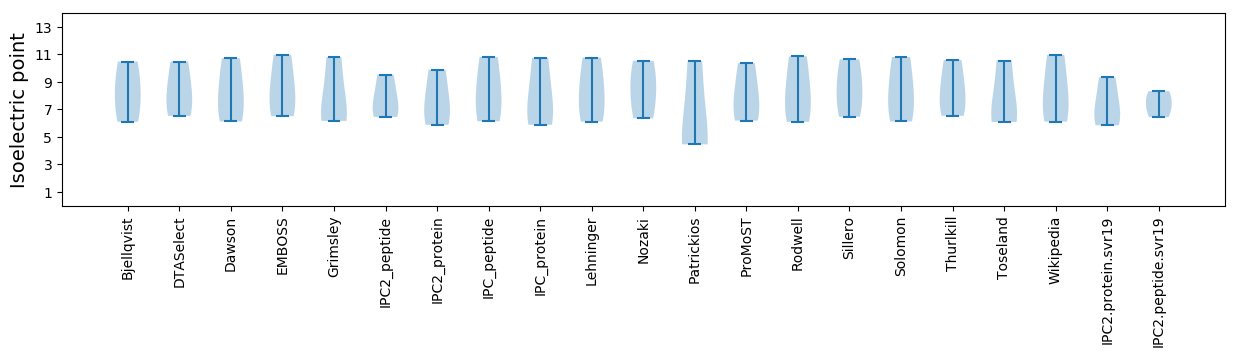
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5UAI8|U5UAI8_9VIRU CA1 OS=Gyrovirus Tu243 OX=1415627 PE=3 SV=1
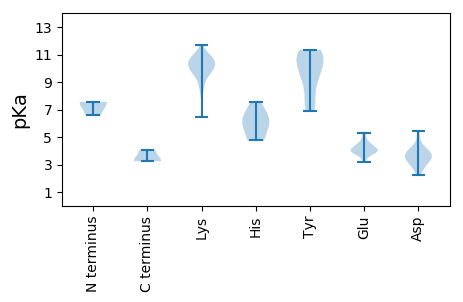
MM1 pKa = 7.48LADD4 pKa = 4.74EE5 pKa = 5.29LPGDD9 pKa = 4.08AQASVQLRR17 pKa = 11.84QFQEE21 pKa = 3.83PLVPVGGEE29 pKa = 3.9ACVGSCYY36 pKa = 10.42GHH38 pKa = 6.34SDD40 pKa = 3.53RR41 pKa = 11.84PISSEE46 pKa = 3.88TSVWWFRR53 pKa = 11.84HH54 pKa = 4.92CRR56 pKa = 11.84DD57 pKa = 2.69WSEE60 pKa = 3.26ARR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.92EE66 pKa = 3.17ILEE69 pKa = 3.92FFKK72 pKa = 10.78VQKK75 pKa = 10.21RR76 pKa = 11.84RR77 pKa = 11.84LPGVGGGLQEE87 pKa = 4.21EE88 pKa = 4.87LL89 pKa = 4.07
MM1 pKa = 7.48LADD4 pKa = 4.74EE5 pKa = 5.29LPGDD9 pKa = 4.08AQASVQLRR17 pKa = 11.84QFQEE21 pKa = 3.83PLVPVGGEE29 pKa = 3.9ACVGSCYY36 pKa = 10.42GHH38 pKa = 6.34SDD40 pKa = 3.53RR41 pKa = 11.84PISSEE46 pKa = 3.88TSVWWFRR53 pKa = 11.84HH54 pKa = 4.92CRR56 pKa = 11.84DD57 pKa = 2.69WSEE60 pKa = 3.26ARR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.92EE66 pKa = 3.17ILEE69 pKa = 3.92FFKK72 pKa = 10.78VQKK75 pKa = 10.21RR76 pKa = 11.84RR77 pKa = 11.84LPGVGGGLQEE87 pKa = 4.21EE88 pKa = 4.87LL89 pKa = 4.07
Molecular weight: 10.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5UAI8|U5UAI8_9VIRU CA1 OS=Gyrovirus Tu243 OX=1415627 PE=3 SV=1
MM1 pKa = 6.57VRR3 pKa = 11.84YY4 pKa = 9.5RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84PWGRR12 pKa = 11.84LYY14 pKa = 10.86KK15 pKa = 9.68WFRR18 pKa = 11.84GGWHH22 pKa = 5.56YY23 pKa = 11.06KK24 pKa = 9.81RR25 pKa = 11.84RR26 pKa = 11.84PKK28 pKa = 10.33GRR30 pKa = 11.84WYY32 pKa = 10.06RR33 pKa = 11.84SSWGRR38 pKa = 11.84KK39 pKa = 6.44RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 6.9HH43 pKa = 4.8HH44 pKa = 7.11RR45 pKa = 11.84YY46 pKa = 9.14RR47 pKa = 11.84RR48 pKa = 11.84GLFGRR53 pKa = 11.84NFKK56 pKa = 10.66RR57 pKa = 11.84SFYY60 pKa = 8.59VQHH63 pKa = 6.88PGSYY67 pKa = 7.84TVRR70 pKa = 11.84RR71 pKa = 11.84TNPYY75 pKa = 7.3NTQRR79 pKa = 11.84LRR81 pKa = 11.84FQGLILVNNAVTQTGTSSVSNMRR104 pKa = 11.84ATAIEE109 pKa = 3.72ISLRR113 pKa = 11.84GLLLAYY119 pKa = 9.79FMNHH123 pKa = 4.75STGGPGWAGDD133 pKa = 3.72SGTKK137 pKa = 10.02VAISPLEE144 pKa = 3.79WWRR147 pKa = 11.84WAYY150 pKa = 9.93IRR152 pKa = 11.84IEE154 pKa = 4.1PCNDD158 pKa = 2.2GRR160 pKa = 11.84VFGMAVPHH168 pKa = 7.52DD169 pKa = 3.9EE170 pKa = 4.53VLYY173 pKa = 11.11NWFCQWQLFKK183 pKa = 10.76HH184 pKa = 5.74VKK186 pKa = 8.69TDD188 pKa = 3.62LRR190 pKa = 11.84VLNTDD195 pKa = 2.66VRR197 pKa = 11.84TSAEE201 pKa = 3.74ARR203 pKa = 11.84IVASLLVQDD212 pKa = 5.43SYY214 pKa = 10.96WQVGNRR220 pKa = 11.84GATQLSQIKK229 pKa = 9.78FPSFLEE235 pKa = 4.05LSAWGAQWSFQPGLQSVSSRR255 pKa = 11.84SFNHH259 pKa = 6.18HH260 pKa = 6.2SFVGTNDD267 pKa = 3.25PMGEE271 pKa = 3.97PWKK274 pKa = 10.7VFCPLDD280 pKa = 4.05NMTQQDD286 pKa = 3.98TQISWVYY293 pKa = 9.29ATLFLAEE300 pKa = 4.45FASVTNKK307 pKa = 9.93QGYY310 pKa = 7.24NTEE313 pKa = 4.13QTSLEE318 pKa = 4.08MQALSDD324 pKa = 3.34TWAVIKK330 pKa = 10.69VRR332 pKa = 11.84SEE334 pKa = 3.46WMLGNQNRR342 pKa = 11.84VKK344 pKa = 11.02SNMVTFTTGEE354 pKa = 4.08PPP356 pKa = 3.24
MM1 pKa = 6.57VRR3 pKa = 11.84YY4 pKa = 9.5RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84PWGRR12 pKa = 11.84LYY14 pKa = 10.86KK15 pKa = 9.68WFRR18 pKa = 11.84GGWHH22 pKa = 5.56YY23 pKa = 11.06KK24 pKa = 9.81RR25 pKa = 11.84RR26 pKa = 11.84PKK28 pKa = 10.33GRR30 pKa = 11.84WYY32 pKa = 10.06RR33 pKa = 11.84SSWGRR38 pKa = 11.84KK39 pKa = 6.44RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 6.9HH43 pKa = 4.8HH44 pKa = 7.11RR45 pKa = 11.84YY46 pKa = 9.14RR47 pKa = 11.84RR48 pKa = 11.84GLFGRR53 pKa = 11.84NFKK56 pKa = 10.66RR57 pKa = 11.84SFYY60 pKa = 8.59VQHH63 pKa = 6.88PGSYY67 pKa = 7.84TVRR70 pKa = 11.84RR71 pKa = 11.84TNPYY75 pKa = 7.3NTQRR79 pKa = 11.84LRR81 pKa = 11.84FQGLILVNNAVTQTGTSSVSNMRR104 pKa = 11.84ATAIEE109 pKa = 3.72ISLRR113 pKa = 11.84GLLLAYY119 pKa = 9.79FMNHH123 pKa = 4.75STGGPGWAGDD133 pKa = 3.72SGTKK137 pKa = 10.02VAISPLEE144 pKa = 3.79WWRR147 pKa = 11.84WAYY150 pKa = 9.93IRR152 pKa = 11.84IEE154 pKa = 4.1PCNDD158 pKa = 2.2GRR160 pKa = 11.84VFGMAVPHH168 pKa = 7.52DD169 pKa = 3.9EE170 pKa = 4.53VLYY173 pKa = 11.11NWFCQWQLFKK183 pKa = 10.76HH184 pKa = 5.74VKK186 pKa = 8.69TDD188 pKa = 3.62LRR190 pKa = 11.84VLNTDD195 pKa = 2.66VRR197 pKa = 11.84TSAEE201 pKa = 3.74ARR203 pKa = 11.84IVASLLVQDD212 pKa = 5.43SYY214 pKa = 10.96WQVGNRR220 pKa = 11.84GATQLSQIKK229 pKa = 9.78FPSFLEE235 pKa = 4.05LSAWGAQWSFQPGLQSVSSRR255 pKa = 11.84SFNHH259 pKa = 6.18HH260 pKa = 6.2SFVGTNDD267 pKa = 3.25PMGEE271 pKa = 3.97PWKK274 pKa = 10.7VFCPLDD280 pKa = 4.05NMTQQDD286 pKa = 3.98TQISWVYY293 pKa = 9.29ATLFLAEE300 pKa = 4.45FASVTNKK307 pKa = 9.93QGYY310 pKa = 7.24NTEE313 pKa = 4.13QTSLEE318 pKa = 4.08MQALSDD324 pKa = 3.34TWAVIKK330 pKa = 10.69VRR332 pKa = 11.84SEE334 pKa = 3.46WMLGNQNRR342 pKa = 11.84VKK344 pKa = 11.02SNMVTFTTGEE354 pKa = 4.08PPP356 pKa = 3.24
Molecular weight: 41.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
676 |
89 |
356 |
225.3 |
25.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.325 ± 0.093 | 1.775 ± 0.738 |
3.698 ± 0.645 | 5.621 ± 1.87 |
4.734 ± 0.233 | 9.911 ± 1.728 |
1.923 ± 0.49 | 3.254 ± 0.514 |
4.734 ± 1.009 | 7.544 ± 0.433 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.923 ± 0.454 | 3.994 ± 1.363 |
4.438 ± 0.357 | 5.178 ± 0.654 |
8.58 ± 1.159 | 8.136 ± 0.082 |
5.917 ± 1.439 | 6.657 ± 0.519 |
3.698 ± 0.995 | 2.959 ± 0.957 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
