
Kwoniella bestiolae CBS 10118
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Kwoniella; Kwoniella bestiolae
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
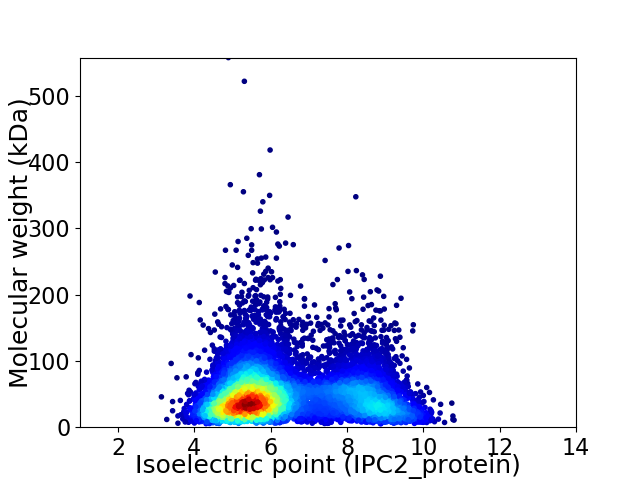
Virtual 2D-PAGE plot for 9131 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9FUC2|A0A1B9FUC2_9TREE Rab-GAP TBC domain-containing protein OS=Kwoniella bestiolae CBS 10118 OX=1296100 GN=I302_08012 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.54SSTTTLTTYY11 pKa = 11.49ALLVLSTIITSVDD24 pKa = 2.68AAGPHH29 pKa = 5.4NRR31 pKa = 11.84FGVGAHH37 pKa = 6.31RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 6.3HH41 pKa = 6.34ARR43 pKa = 11.84AAATSTEE50 pKa = 4.02LSTPVQSSEE59 pKa = 4.03QKK61 pKa = 10.2RR62 pKa = 11.84DD63 pKa = 3.43EE64 pKa = 4.76CEE66 pKa = 3.58LGAWKK71 pKa = 10.53CVGSVLQRR79 pKa = 11.84CYY81 pKa = 10.44NDD83 pKa = 2.64QWNFVTNCTGTDD95 pKa = 4.03IICSDD100 pKa = 3.33GLYY103 pKa = 7.57TTGCVWTWSVEE114 pKa = 4.19QNDD117 pKa = 4.55DD118 pKa = 3.82ANEE121 pKa = 4.24SATSSPPFTSSITVTPTASEE141 pKa = 4.41SSASASTASITNANLAVEE159 pKa = 4.61PTATATADD167 pKa = 3.39ANEE170 pKa = 4.56DD171 pKa = 3.95EE172 pKa = 4.54EE173 pKa = 6.35CEE175 pKa = 5.39DD176 pKa = 4.65EE177 pKa = 6.43DD178 pKa = 4.9EE179 pKa = 5.92QDD181 pKa = 3.97DD182 pKa = 4.61QEE184 pKa = 5.66DD185 pKa = 4.23DD186 pKa = 3.57EE187 pKa = 6.39DD188 pKa = 4.51EE189 pKa = 4.97GEE191 pKa = 4.47EE192 pKa = 4.25YY193 pKa = 10.79CDD195 pKa = 6.01DD196 pKa = 4.4EE197 pKa = 7.72DD198 pKa = 5.92DD199 pKa = 4.71GAWDD203 pKa = 4.25EE204 pKa = 4.79YY205 pKa = 11.08YY206 pKa = 11.12SSATAVVTATPTSAASVVTAAPTSAIGGDD235 pKa = 4.11LAPSDD240 pKa = 5.29DD241 pKa = 4.98DD242 pKa = 6.52DD243 pKa = 5.81EE244 pKa = 7.73GDD246 pKa = 4.58DD247 pKa = 4.15DD248 pKa = 6.34DD249 pKa = 6.15EE250 pKa = 5.11EE251 pKa = 4.76EE252 pKa = 4.24EE253 pKa = 5.54CEE255 pKa = 5.81DD256 pKa = 5.5DD257 pKa = 6.6DD258 pKa = 6.85SDD260 pKa = 6.06DD261 pKa = 4.63YY262 pKa = 12.21DD263 pKa = 4.17DD264 pKa = 5.8ASPTTTSSASTSATSIIGGQLWAGHH289 pKa = 6.4GDD291 pKa = 3.29ASSASTHH298 pKa = 6.44DD299 pKa = 3.35WYY301 pKa = 11.7SWGTQATATTEE312 pKa = 4.15DD313 pKa = 3.81WNTWDD318 pKa = 3.44QATSTSEE325 pKa = 4.42DD326 pKa = 3.37WNTWAQATASASASATSSRR345 pKa = 11.84GHH347 pKa = 6.38GKK349 pKa = 10.69GKK351 pKa = 8.73GHH353 pKa = 7.07KK354 pKa = 9.9SSSADD359 pKa = 3.04WAASTSPSWDD369 pKa = 3.04QSGDD373 pKa = 3.44VSSTTDD379 pKa = 2.83EE380 pKa = 4.22WSTATTTNSWDD391 pKa = 3.56DD392 pKa = 3.28WSGQATASSAAWGQSGKK409 pKa = 11.0GSATASASSSAYY421 pKa = 9.31SAKK424 pKa = 9.9PSKK427 pKa = 10.66GSNHH431 pKa = 4.97TTGSPSSSYY440 pKa = 11.14SSAPHH445 pKa = 4.59YY446 pKa = 10.53VIYY449 pKa = 10.55SDD451 pKa = 3.38YY452 pKa = 10.6WLFEE456 pKa = 4.13MPSVDD461 pKa = 3.51VLSNFNRR468 pKa = 11.84FILAFWMTDD477 pKa = 2.66RR478 pKa = 11.84GAVDD482 pKa = 4.07NAQFWEE488 pKa = 4.21QLDD491 pKa = 3.81STTRR495 pKa = 11.84QQVITDD501 pKa = 3.3YY502 pKa = 10.81HH503 pKa = 5.65NAGIALMVSAFGSTDD518 pKa = 3.12APTSNGADD526 pKa = 3.51PTQTAQALAKK536 pKa = 8.78WVKK539 pKa = 10.57DD540 pKa = 3.6YY541 pKa = 11.68GLDD544 pKa = 3.9GVDD547 pKa = 4.03IDD549 pKa = 4.51YY550 pKa = 11.39EE551 pKa = 4.42DD552 pKa = 3.79MPAMNSAQAVAWIVTFQKK570 pKa = 10.34EE571 pKa = 4.22LRR573 pKa = 11.84RR574 pKa = 11.84LLPSPYY580 pKa = 9.07IISHH584 pKa = 6.48APVAPWFTSANDD596 pKa = 3.86YY597 pKa = 11.02SDD599 pKa = 3.34KK600 pKa = 11.11AYY602 pKa = 10.99VAIHH606 pKa = 6.01EE607 pKa = 4.3QTGDD611 pKa = 3.67GIDD614 pKa = 3.91FYY616 pKa = 11.62NIQFYY621 pKa = 10.48NQGSGVYY628 pKa = 8.56EE629 pKa = 4.02DD630 pKa = 5.17CYY632 pKa = 11.77SLLFDD637 pKa = 5.61SGNDD641 pKa = 3.41WPSTSVFEE649 pKa = 4.69INSSAGLPLDD659 pKa = 4.54KK660 pKa = 10.61IVIGKK665 pKa = 9.09PLDD668 pKa = 3.58EE669 pKa = 4.45GAAANGFIDD678 pKa = 4.33ASLLAKK684 pKa = 10.21CVSQAQAKK692 pKa = 8.22GWNAGVMFWEE702 pKa = 4.49WTTVSPAVPSS712 pKa = 3.68
MM1 pKa = 7.43KK2 pKa = 10.54SSTTTLTTYY11 pKa = 11.49ALLVLSTIITSVDD24 pKa = 2.68AAGPHH29 pKa = 5.4NRR31 pKa = 11.84FGVGAHH37 pKa = 6.31RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 6.3HH41 pKa = 6.34ARR43 pKa = 11.84AAATSTEE50 pKa = 4.02LSTPVQSSEE59 pKa = 4.03QKK61 pKa = 10.2RR62 pKa = 11.84DD63 pKa = 3.43EE64 pKa = 4.76CEE66 pKa = 3.58LGAWKK71 pKa = 10.53CVGSVLQRR79 pKa = 11.84CYY81 pKa = 10.44NDD83 pKa = 2.64QWNFVTNCTGTDD95 pKa = 4.03IICSDD100 pKa = 3.33GLYY103 pKa = 7.57TTGCVWTWSVEE114 pKa = 4.19QNDD117 pKa = 4.55DD118 pKa = 3.82ANEE121 pKa = 4.24SATSSPPFTSSITVTPTASEE141 pKa = 4.41SSASASTASITNANLAVEE159 pKa = 4.61PTATATADD167 pKa = 3.39ANEE170 pKa = 4.56DD171 pKa = 3.95EE172 pKa = 4.54EE173 pKa = 6.35CEE175 pKa = 5.39DD176 pKa = 4.65EE177 pKa = 6.43DD178 pKa = 4.9EE179 pKa = 5.92QDD181 pKa = 3.97DD182 pKa = 4.61QEE184 pKa = 5.66DD185 pKa = 4.23DD186 pKa = 3.57EE187 pKa = 6.39DD188 pKa = 4.51EE189 pKa = 4.97GEE191 pKa = 4.47EE192 pKa = 4.25YY193 pKa = 10.79CDD195 pKa = 6.01DD196 pKa = 4.4EE197 pKa = 7.72DD198 pKa = 5.92DD199 pKa = 4.71GAWDD203 pKa = 4.25EE204 pKa = 4.79YY205 pKa = 11.08YY206 pKa = 11.12SSATAVVTATPTSAASVVTAAPTSAIGGDD235 pKa = 4.11LAPSDD240 pKa = 5.29DD241 pKa = 4.98DD242 pKa = 6.52DD243 pKa = 5.81EE244 pKa = 7.73GDD246 pKa = 4.58DD247 pKa = 4.15DD248 pKa = 6.34DD249 pKa = 6.15EE250 pKa = 5.11EE251 pKa = 4.76EE252 pKa = 4.24EE253 pKa = 5.54CEE255 pKa = 5.81DD256 pKa = 5.5DD257 pKa = 6.6DD258 pKa = 6.85SDD260 pKa = 6.06DD261 pKa = 4.63YY262 pKa = 12.21DD263 pKa = 4.17DD264 pKa = 5.8ASPTTTSSASTSATSIIGGQLWAGHH289 pKa = 6.4GDD291 pKa = 3.29ASSASTHH298 pKa = 6.44DD299 pKa = 3.35WYY301 pKa = 11.7SWGTQATATTEE312 pKa = 4.15DD313 pKa = 3.81WNTWDD318 pKa = 3.44QATSTSEE325 pKa = 4.42DD326 pKa = 3.37WNTWAQATASASASATSSRR345 pKa = 11.84GHH347 pKa = 6.38GKK349 pKa = 10.69GKK351 pKa = 8.73GHH353 pKa = 7.07KK354 pKa = 9.9SSSADD359 pKa = 3.04WAASTSPSWDD369 pKa = 3.04QSGDD373 pKa = 3.44VSSTTDD379 pKa = 2.83EE380 pKa = 4.22WSTATTTNSWDD391 pKa = 3.56DD392 pKa = 3.28WSGQATASSAAWGQSGKK409 pKa = 11.0GSATASASSSAYY421 pKa = 9.31SAKK424 pKa = 9.9PSKK427 pKa = 10.66GSNHH431 pKa = 4.97TTGSPSSSYY440 pKa = 11.14SSAPHH445 pKa = 4.59YY446 pKa = 10.53VIYY449 pKa = 10.55SDD451 pKa = 3.38YY452 pKa = 10.6WLFEE456 pKa = 4.13MPSVDD461 pKa = 3.51VLSNFNRR468 pKa = 11.84FILAFWMTDD477 pKa = 2.66RR478 pKa = 11.84GAVDD482 pKa = 4.07NAQFWEE488 pKa = 4.21QLDD491 pKa = 3.81STTRR495 pKa = 11.84QQVITDD501 pKa = 3.3YY502 pKa = 10.81HH503 pKa = 5.65NAGIALMVSAFGSTDD518 pKa = 3.12APTSNGADD526 pKa = 3.51PTQTAQALAKK536 pKa = 8.78WVKK539 pKa = 10.57DD540 pKa = 3.6YY541 pKa = 11.68GLDD544 pKa = 3.9GVDD547 pKa = 4.03IDD549 pKa = 4.51YY550 pKa = 11.39EE551 pKa = 4.42DD552 pKa = 3.79MPAMNSAQAVAWIVTFQKK570 pKa = 10.34EE571 pKa = 4.22LRR573 pKa = 11.84RR574 pKa = 11.84LLPSPYY580 pKa = 9.07IISHH584 pKa = 6.48APVAPWFTSANDD596 pKa = 3.86YY597 pKa = 11.02SDD599 pKa = 3.34KK600 pKa = 11.11AYY602 pKa = 10.99VAIHH606 pKa = 6.01EE607 pKa = 4.3QTGDD611 pKa = 3.67GIDD614 pKa = 3.91FYY616 pKa = 11.62NIQFYY621 pKa = 10.48NQGSGVYY628 pKa = 8.56EE629 pKa = 4.02DD630 pKa = 5.17CYY632 pKa = 11.77SLLFDD637 pKa = 5.61SGNDD641 pKa = 3.41WPSTSVFEE649 pKa = 4.69INSSAGLPLDD659 pKa = 4.54KK660 pKa = 10.61IVIGKK665 pKa = 9.09PLDD668 pKa = 3.58EE669 pKa = 4.45GAAANGFIDD678 pKa = 4.33ASLLAKK684 pKa = 10.21CVSQAQAKK692 pKa = 8.22GWNAGVMFWEE702 pKa = 4.49WTTVSPAVPSS712 pKa = 3.68
Molecular weight: 75.92 kDa
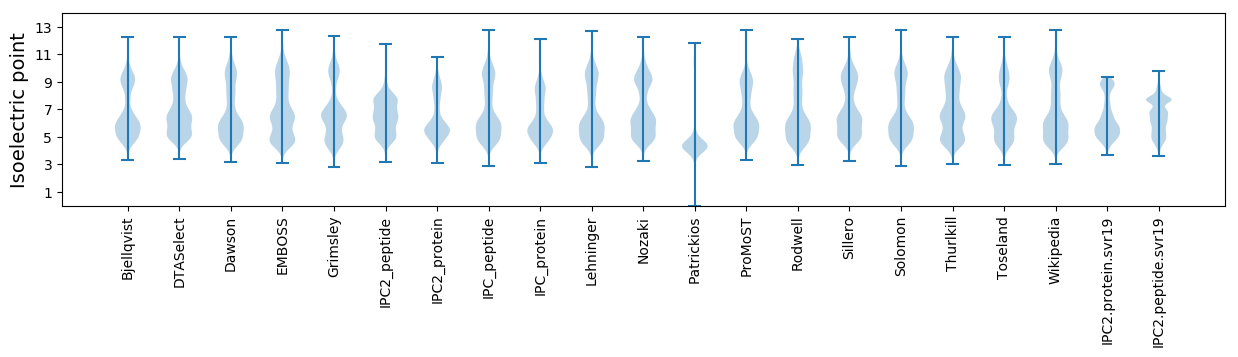
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9GEZ9|A0A1B9GEZ9_9TREE Flavin mononucleotide kinase 1 OS=Kwoniella bestiolae CBS 10118 OX=1296100 GN=I302_01056 PE=3 SV=1
MM1 pKa = 7.85PSLNQLCQNCGLPRR15 pKa = 11.84ASPPPPLDD23 pKa = 3.27TMFEE27 pKa = 4.18EE28 pKa = 4.77DD29 pKa = 4.18FEE31 pKa = 5.19RR32 pKa = 11.84CFICQQPCKK41 pKa = 10.49GLYY44 pKa = 9.88CSSEE48 pKa = 3.78CRR50 pKa = 11.84LRR52 pKa = 11.84DD53 pKa = 3.24QGTPSPAVRR62 pKa = 11.84AAHH65 pKa = 6.13GPVKK69 pKa = 9.77ITSQLPAALSPLVRR83 pKa = 11.84PTQHH87 pKa = 7.16PGRR90 pKa = 11.84SPRR93 pKa = 11.84VLAQNRR99 pKa = 11.84GSSSISSGSSSVSSSPLQSPQTNPSEE125 pKa = 4.15VDD127 pKa = 3.24SPKK130 pKa = 10.24RR131 pKa = 11.84DD132 pKa = 3.71NFDD135 pKa = 4.39LPPPAYY141 pKa = 7.38PTKK144 pKa = 10.7QFGAPASVPMKK155 pKa = 10.38IPALASRR162 pKa = 11.84PSPLVAPSQTPGSVGSTVYY181 pKa = 10.23PAGASIDD188 pKa = 3.47TLRR191 pKa = 11.84YY192 pKa = 8.36GRR194 pKa = 11.84KK195 pKa = 8.05PSAVNSVISPNALIPRR211 pKa = 11.84CACGKK216 pKa = 9.03PANHH220 pKa = 7.4KK221 pKa = 10.24NRR223 pKa = 11.84ASSKK227 pKa = 10.73DD228 pKa = 3.42RR229 pKa = 11.84ADD231 pKa = 4.9LIDD234 pKa = 3.86SGFSRR239 pKa = 11.84LSLGPSLVTAQEE251 pKa = 4.14EE252 pKa = 4.13PLPRR256 pKa = 11.84STRR259 pKa = 11.84IVSEE263 pKa = 4.0SAIPPFISGTPGRR276 pKa = 11.84KK277 pKa = 7.14TVPLGIPSSPQVAPSTSLLSRR298 pKa = 11.84SRR300 pKa = 11.84SDD302 pKa = 4.68PIPGSPQTQRR312 pKa = 11.84KK313 pKa = 9.21LIPQVPAPVITNVITPSHH331 pKa = 6.68RR332 pKa = 11.84EE333 pKa = 4.02SNGMPLSPIVPPATRR348 pKa = 11.84PGRR351 pKa = 11.84NRR353 pKa = 11.84NLEE356 pKa = 4.07VNMDD360 pKa = 3.43SPRR363 pKa = 11.84RR364 pKa = 11.84GRR366 pKa = 11.84SRR368 pKa = 11.84EE369 pKa = 3.92RR370 pKa = 11.84QEE372 pKa = 3.92HH373 pKa = 6.26HH374 pKa = 6.37VGQMTSNFGGPADD387 pKa = 4.34RR388 pKa = 11.84EE389 pKa = 4.26QAPSRR394 pKa = 11.84SRR396 pKa = 11.84TRR398 pKa = 11.84RR399 pKa = 11.84RR400 pKa = 11.84SDD402 pKa = 2.76SRR404 pKa = 11.84EE405 pKa = 3.7RR406 pKa = 11.84EE407 pKa = 3.9RR408 pKa = 11.84EE409 pKa = 3.58RR410 pKa = 11.84GRR412 pKa = 11.84GRR414 pKa = 11.84DD415 pKa = 3.08SRR417 pKa = 11.84EE418 pKa = 3.54RR419 pKa = 11.84EE420 pKa = 3.77RR421 pKa = 11.84EE422 pKa = 3.76RR423 pKa = 11.84DD424 pKa = 3.64HH425 pKa = 7.29SGHH428 pKa = 6.39TSPIKK433 pKa = 9.4QTQTPQILPSWSRR446 pKa = 11.84RR447 pKa = 11.84ASEE450 pKa = 3.8ATADD454 pKa = 3.7RR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 10.53VLGEE461 pKa = 3.59VAPAMRR467 pKa = 11.84RR468 pKa = 11.84TASNGKK474 pKa = 9.78KK475 pKa = 10.2SPTYY479 pKa = 10.4DD480 pKa = 3.4EE481 pKa = 4.76EE482 pKa = 4.07EE483 pKa = 4.13KK484 pKa = 10.53QRR486 pKa = 11.84KK487 pKa = 8.54KK488 pKa = 11.23DD489 pKa = 3.68EE490 pKa = 4.07INRR493 pKa = 11.84ASKK496 pKa = 10.61QLGQVFGVAAGG507 pKa = 3.38
MM1 pKa = 7.85PSLNQLCQNCGLPRR15 pKa = 11.84ASPPPPLDD23 pKa = 3.27TMFEE27 pKa = 4.18EE28 pKa = 4.77DD29 pKa = 4.18FEE31 pKa = 5.19RR32 pKa = 11.84CFICQQPCKK41 pKa = 10.49GLYY44 pKa = 9.88CSSEE48 pKa = 3.78CRR50 pKa = 11.84LRR52 pKa = 11.84DD53 pKa = 3.24QGTPSPAVRR62 pKa = 11.84AAHH65 pKa = 6.13GPVKK69 pKa = 9.77ITSQLPAALSPLVRR83 pKa = 11.84PTQHH87 pKa = 7.16PGRR90 pKa = 11.84SPRR93 pKa = 11.84VLAQNRR99 pKa = 11.84GSSSISSGSSSVSSSPLQSPQTNPSEE125 pKa = 4.15VDD127 pKa = 3.24SPKK130 pKa = 10.24RR131 pKa = 11.84DD132 pKa = 3.71NFDD135 pKa = 4.39LPPPAYY141 pKa = 7.38PTKK144 pKa = 10.7QFGAPASVPMKK155 pKa = 10.38IPALASRR162 pKa = 11.84PSPLVAPSQTPGSVGSTVYY181 pKa = 10.23PAGASIDD188 pKa = 3.47TLRR191 pKa = 11.84YY192 pKa = 8.36GRR194 pKa = 11.84KK195 pKa = 8.05PSAVNSVISPNALIPRR211 pKa = 11.84CACGKK216 pKa = 9.03PANHH220 pKa = 7.4KK221 pKa = 10.24NRR223 pKa = 11.84ASSKK227 pKa = 10.73DD228 pKa = 3.42RR229 pKa = 11.84ADD231 pKa = 4.9LIDD234 pKa = 3.86SGFSRR239 pKa = 11.84LSLGPSLVTAQEE251 pKa = 4.14EE252 pKa = 4.13PLPRR256 pKa = 11.84STRR259 pKa = 11.84IVSEE263 pKa = 4.0SAIPPFISGTPGRR276 pKa = 11.84KK277 pKa = 7.14TVPLGIPSSPQVAPSTSLLSRR298 pKa = 11.84SRR300 pKa = 11.84SDD302 pKa = 4.68PIPGSPQTQRR312 pKa = 11.84KK313 pKa = 9.21LIPQVPAPVITNVITPSHH331 pKa = 6.68RR332 pKa = 11.84EE333 pKa = 4.02SNGMPLSPIVPPATRR348 pKa = 11.84PGRR351 pKa = 11.84NRR353 pKa = 11.84NLEE356 pKa = 4.07VNMDD360 pKa = 3.43SPRR363 pKa = 11.84RR364 pKa = 11.84GRR366 pKa = 11.84SRR368 pKa = 11.84EE369 pKa = 3.92RR370 pKa = 11.84QEE372 pKa = 3.92HH373 pKa = 6.26HH374 pKa = 6.37VGQMTSNFGGPADD387 pKa = 4.34RR388 pKa = 11.84EE389 pKa = 4.26QAPSRR394 pKa = 11.84SRR396 pKa = 11.84TRR398 pKa = 11.84RR399 pKa = 11.84RR400 pKa = 11.84SDD402 pKa = 2.76SRR404 pKa = 11.84EE405 pKa = 3.7RR406 pKa = 11.84EE407 pKa = 3.9RR408 pKa = 11.84EE409 pKa = 3.58RR410 pKa = 11.84GRR412 pKa = 11.84GRR414 pKa = 11.84DD415 pKa = 3.08SRR417 pKa = 11.84EE418 pKa = 3.54RR419 pKa = 11.84EE420 pKa = 3.77RR421 pKa = 11.84EE422 pKa = 3.76RR423 pKa = 11.84DD424 pKa = 3.64HH425 pKa = 7.29SGHH428 pKa = 6.39TSPIKK433 pKa = 9.4QTQTPQILPSWSRR446 pKa = 11.84RR447 pKa = 11.84ASEE450 pKa = 3.8ATADD454 pKa = 3.7RR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 10.53VLGEE461 pKa = 3.59VAPAMRR467 pKa = 11.84RR468 pKa = 11.84TASNGKK474 pKa = 9.78KK475 pKa = 10.2SPTYY479 pKa = 10.4DD480 pKa = 3.4EE481 pKa = 4.76EE482 pKa = 4.07EE483 pKa = 4.13KK484 pKa = 10.53QRR486 pKa = 11.84KK487 pKa = 8.54KK488 pKa = 11.23DD489 pKa = 3.68EE490 pKa = 4.07INRR493 pKa = 11.84ASKK496 pKa = 10.61QLGQVFGVAAGG507 pKa = 3.38
Molecular weight: 54.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4442483 |
49 |
5016 |
486.5 |
53.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.382 ± 0.029 | 1.003 ± 0.009 |
5.562 ± 0.019 | 6.529 ± 0.027 |
3.335 ± 0.015 | 7.258 ± 0.023 |
2.325 ± 0.011 | 5.119 ± 0.017 |
5.24 ± 0.024 | 8.764 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.009 | 3.781 ± 0.016 |
6.669 ± 0.031 | 3.883 ± 0.023 |
5.791 ± 0.022 | 9.442 ± 0.033 |
6.101 ± 0.017 | 5.732 ± 0.018 |
1.377 ± 0.009 | 2.635 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
