
Leptonychotes weddellii papillomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
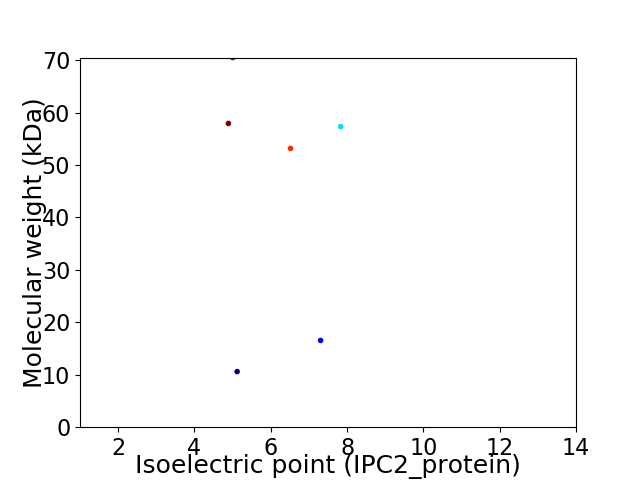
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
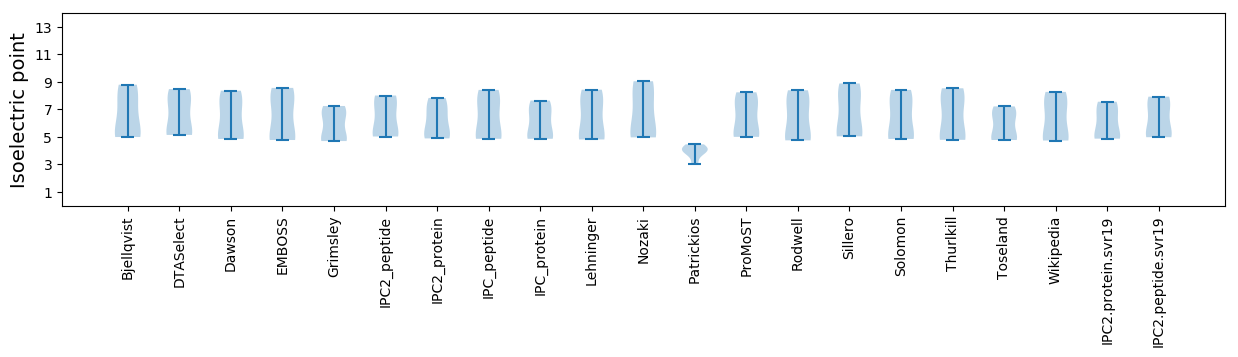
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I8B2R1|A0A2I8B2R1_9PAPI Minor capsid protein L2 OS=Leptonychotes weddellii papillomavirus 4 OX=2077305 GN=L2 PE=3 SV=1
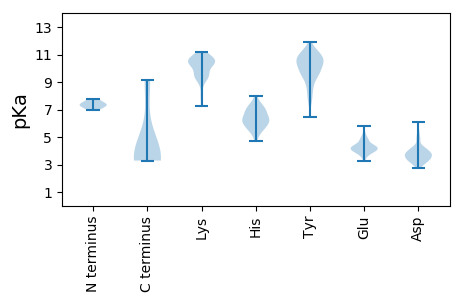
MM1 pKa = 7.4IGKK4 pKa = 9.27EE5 pKa = 3.72PTLRR9 pKa = 11.84DD10 pKa = 3.17IVLEE14 pKa = 4.03EE15 pKa = 4.54TPCPADD21 pKa = 3.91LYY23 pKa = 9.64CNEE26 pKa = 4.78RR27 pKa = 11.84IEE29 pKa = 4.33EE30 pKa = 4.23AEE32 pKa = 4.06QNISEE37 pKa = 4.37GNHH40 pKa = 6.34DD41 pKa = 3.96AFKK44 pKa = 11.22VLVTCGHH51 pKa = 6.96CDD53 pKa = 2.98RR54 pKa = 11.84PVRR57 pKa = 11.84LVILASRR64 pKa = 11.84DD65 pKa = 3.51QVRR68 pKa = 11.84VLHH71 pKa = 6.51RR72 pKa = 11.84LLLEE76 pKa = 4.3GLCFVCPYY84 pKa = 9.85CCRR87 pKa = 11.84EE88 pKa = 3.37RR89 pKa = 11.84DD90 pKa = 3.75YY91 pKa = 11.94NN92 pKa = 3.67
MM1 pKa = 7.4IGKK4 pKa = 9.27EE5 pKa = 3.72PTLRR9 pKa = 11.84DD10 pKa = 3.17IVLEE14 pKa = 4.03EE15 pKa = 4.54TPCPADD21 pKa = 3.91LYY23 pKa = 9.64CNEE26 pKa = 4.78RR27 pKa = 11.84IEE29 pKa = 4.33EE30 pKa = 4.23AEE32 pKa = 4.06QNISEE37 pKa = 4.37GNHH40 pKa = 6.34DD41 pKa = 3.96AFKK44 pKa = 11.22VLVTCGHH51 pKa = 6.96CDD53 pKa = 2.98RR54 pKa = 11.84PVRR57 pKa = 11.84LVILASRR64 pKa = 11.84DD65 pKa = 3.51QVRR68 pKa = 11.84VLHH71 pKa = 6.51RR72 pKa = 11.84LLLEE76 pKa = 4.3GLCFVCPYY84 pKa = 9.85CCRR87 pKa = 11.84EE88 pKa = 3.37RR89 pKa = 11.84DD90 pKa = 3.75YY91 pKa = 11.94NN92 pKa = 3.67
Molecular weight: 10.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I8B2R5|A0A2I8B2R5_9PAPI Regulatory protein E2 OS=Leptonychotes weddellii papillomavirus 4 OX=2077305 GN=E2 PE=3 SV=1
MM1 pKa = 6.97EE2 pKa = 4.63TLSRR6 pKa = 11.84RR7 pKa = 11.84LDD9 pKa = 3.59ALQEE13 pKa = 4.08EE14 pKa = 5.04LLNLYY19 pKa = 8.49EE20 pKa = 4.81KK21 pKa = 10.77SSEE24 pKa = 4.02QLEE27 pKa = 4.33HH28 pKa = 7.28QILHH32 pKa = 5.49WQLLRR37 pKa = 11.84QEE39 pKa = 4.53NVLLHH44 pKa = 5.05YY45 pKa = 10.25AKK47 pKa = 10.5KK48 pKa = 10.51RR49 pKa = 11.84GVCRR53 pKa = 11.84LGYY56 pKa = 9.47QAVPAAVVTQEE67 pKa = 4.03KK68 pKa = 10.09AKK70 pKa = 10.43QAIEE74 pKa = 4.07MEE76 pKa = 4.39LVLTSLSKK84 pKa = 10.75SAFSKK89 pKa = 10.61EE90 pKa = 3.76PWTLTDD96 pKa = 3.13TSRR99 pKa = 11.84DD100 pKa = 3.32MWCAEE105 pKa = 3.74PRR107 pKa = 11.84HH108 pKa = 5.94CFKK111 pKa = 11.02KK112 pKa = 10.54GGSQVEE118 pKa = 4.75VVFGACPSDD127 pKa = 3.76SMLYY131 pKa = 7.99TCWGSVYY138 pKa = 10.69YY139 pKa = 10.47QDD141 pKa = 6.13ADD143 pKa = 4.21DD144 pKa = 4.2VWHH147 pKa = 6.37KK148 pKa = 9.77TSSTVCHH155 pKa = 6.32AGVFFMEE162 pKa = 5.53GSQAKK167 pKa = 9.92YY168 pKa = 9.97YY169 pKa = 11.14VDD171 pKa = 4.9FSSEE175 pKa = 3.58AARR178 pKa = 11.84YY179 pKa = 7.54GASDD183 pKa = 3.03WTVIHH188 pKa = 5.95KK189 pKa = 7.99TAVYY193 pKa = 10.3SSVAVTSSEE202 pKa = 4.14CGEE205 pKa = 4.02AAAGSDD211 pKa = 3.33QRR213 pKa = 11.84RR214 pKa = 11.84AGVVRR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84GRR223 pKa = 11.84PPKK226 pKa = 10.23KK227 pKa = 9.53YY228 pKa = 10.38GRR230 pKa = 11.84AASPLPTEE238 pKa = 4.23TQEE241 pKa = 4.23TVTDD245 pKa = 4.27SVAPGYY251 pKa = 9.5PGLPGPPGLPGPPGPKK267 pKa = 9.44RR268 pKa = 11.84PCTGWGGPTTSGASPGACEE287 pKa = 4.18PSITAASATDD297 pKa = 3.69PTGSRR302 pKa = 11.84LPPLTVSGQLGRR314 pKa = 11.84GQGEE318 pKa = 4.31PEE320 pKa = 3.88TRR322 pKa = 11.84STFWQQEE329 pKa = 4.31EE330 pKa = 5.14GEE332 pKa = 4.45HH333 pKa = 5.57TRR335 pKa = 11.84SEE337 pKa = 4.44PEE339 pKa = 3.67STPPRR344 pKa = 11.84EE345 pKa = 4.59GYY347 pKa = 10.93ANPRR351 pKa = 11.84PDD353 pKa = 3.72SSTPDD358 pKa = 3.27PVVARR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84SPGLGRR371 pKa = 11.84SPVWWPPCALPGEE384 pKa = 4.46IPPPVARR391 pKa = 11.84HH392 pKa = 5.17NSEE395 pKa = 3.96SPRR398 pKa = 11.84CEE400 pKa = 3.88DD401 pKa = 3.24SPPVLEE407 pKa = 4.85EE408 pKa = 4.06EE409 pKa = 4.45ACEE412 pKa = 4.09GHH414 pKa = 6.43SPTSSKK420 pKa = 10.48RR421 pKa = 11.84KK422 pKa = 9.29QPEE425 pKa = 3.42QTKK428 pKa = 9.61RR429 pKa = 11.84SRR431 pKa = 11.84RR432 pKa = 11.84PRR434 pKa = 11.84AVLQPAGSSPVVLLKK449 pKa = 10.92GGCNQLKK456 pKa = 9.65CLRR459 pKa = 11.84YY460 pKa = 9.63RR461 pKa = 11.84LRR463 pKa = 11.84TKK465 pKa = 9.39HH466 pKa = 6.05KK467 pKa = 8.2KK468 pKa = 7.27WVRR471 pKa = 11.84LVSTTFFWVGNDD483 pKa = 2.98SVNRR487 pKa = 11.84LGRR490 pKa = 11.84ARR492 pKa = 11.84ILVTFRR498 pKa = 11.84SHH500 pKa = 6.87AACEE504 pKa = 3.99EE505 pKa = 4.1FLNSGNVPQGVEE517 pKa = 3.3AALIHH522 pKa = 6.12GLL524 pKa = 3.68
MM1 pKa = 6.97EE2 pKa = 4.63TLSRR6 pKa = 11.84RR7 pKa = 11.84LDD9 pKa = 3.59ALQEE13 pKa = 4.08EE14 pKa = 5.04LLNLYY19 pKa = 8.49EE20 pKa = 4.81KK21 pKa = 10.77SSEE24 pKa = 4.02QLEE27 pKa = 4.33HH28 pKa = 7.28QILHH32 pKa = 5.49WQLLRR37 pKa = 11.84QEE39 pKa = 4.53NVLLHH44 pKa = 5.05YY45 pKa = 10.25AKK47 pKa = 10.5KK48 pKa = 10.51RR49 pKa = 11.84GVCRR53 pKa = 11.84LGYY56 pKa = 9.47QAVPAAVVTQEE67 pKa = 4.03KK68 pKa = 10.09AKK70 pKa = 10.43QAIEE74 pKa = 4.07MEE76 pKa = 4.39LVLTSLSKK84 pKa = 10.75SAFSKK89 pKa = 10.61EE90 pKa = 3.76PWTLTDD96 pKa = 3.13TSRR99 pKa = 11.84DD100 pKa = 3.32MWCAEE105 pKa = 3.74PRR107 pKa = 11.84HH108 pKa = 5.94CFKK111 pKa = 11.02KK112 pKa = 10.54GGSQVEE118 pKa = 4.75VVFGACPSDD127 pKa = 3.76SMLYY131 pKa = 7.99TCWGSVYY138 pKa = 10.69YY139 pKa = 10.47QDD141 pKa = 6.13ADD143 pKa = 4.21DD144 pKa = 4.2VWHH147 pKa = 6.37KK148 pKa = 9.77TSSTVCHH155 pKa = 6.32AGVFFMEE162 pKa = 5.53GSQAKK167 pKa = 9.92YY168 pKa = 9.97YY169 pKa = 11.14VDD171 pKa = 4.9FSSEE175 pKa = 3.58AARR178 pKa = 11.84YY179 pKa = 7.54GASDD183 pKa = 3.03WTVIHH188 pKa = 5.95KK189 pKa = 7.99TAVYY193 pKa = 10.3SSVAVTSSEE202 pKa = 4.14CGEE205 pKa = 4.02AAAGSDD211 pKa = 3.33QRR213 pKa = 11.84RR214 pKa = 11.84AGVVRR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84GRR223 pKa = 11.84PPKK226 pKa = 10.23KK227 pKa = 9.53YY228 pKa = 10.38GRR230 pKa = 11.84AASPLPTEE238 pKa = 4.23TQEE241 pKa = 4.23TVTDD245 pKa = 4.27SVAPGYY251 pKa = 9.5PGLPGPPGLPGPPGPKK267 pKa = 9.44RR268 pKa = 11.84PCTGWGGPTTSGASPGACEE287 pKa = 4.18PSITAASATDD297 pKa = 3.69PTGSRR302 pKa = 11.84LPPLTVSGQLGRR314 pKa = 11.84GQGEE318 pKa = 4.31PEE320 pKa = 3.88TRR322 pKa = 11.84STFWQQEE329 pKa = 4.31EE330 pKa = 5.14GEE332 pKa = 4.45HH333 pKa = 5.57TRR335 pKa = 11.84SEE337 pKa = 4.44PEE339 pKa = 3.67STPPRR344 pKa = 11.84EE345 pKa = 4.59GYY347 pKa = 10.93ANPRR351 pKa = 11.84PDD353 pKa = 3.72SSTPDD358 pKa = 3.27PVVARR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84SPGLGRR371 pKa = 11.84SPVWWPPCALPGEE384 pKa = 4.46IPPPVARR391 pKa = 11.84HH392 pKa = 5.17NSEE395 pKa = 3.96SPRR398 pKa = 11.84CEE400 pKa = 3.88DD401 pKa = 3.24SPPVLEE407 pKa = 4.85EE408 pKa = 4.06EE409 pKa = 4.45ACEE412 pKa = 4.09GHH414 pKa = 6.43SPTSSKK420 pKa = 10.48RR421 pKa = 11.84KK422 pKa = 9.29QPEE425 pKa = 3.42QTKK428 pKa = 9.61RR429 pKa = 11.84SRR431 pKa = 11.84RR432 pKa = 11.84PRR434 pKa = 11.84AVLQPAGSSPVVLLKK449 pKa = 10.92GGCNQLKK456 pKa = 9.65CLRR459 pKa = 11.84YY460 pKa = 9.63RR461 pKa = 11.84LRR463 pKa = 11.84TKK465 pKa = 9.39HH466 pKa = 6.05KK467 pKa = 8.2KK468 pKa = 7.27WVRR471 pKa = 11.84LVSTTFFWVGNDD483 pKa = 2.98SVNRR487 pKa = 11.84LGRR490 pKa = 11.84ARR492 pKa = 11.84ILVTFRR498 pKa = 11.84SHH500 pKa = 6.87AACEE504 pKa = 3.99EE505 pKa = 4.1FLNSGNVPQGVEE517 pKa = 3.3AALIHH522 pKa = 6.12GLL524 pKa = 3.68
Molecular weight: 57.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2403 |
92 |
628 |
400.5 |
44.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.825 ± 0.546 | 2.622 ± 0.702 |
6.117 ± 0.78 | 6.409 ± 0.506 |
3.829 ± 0.45 | 7.74 ± 1.001 |
2.164 ± 0.209 | 2.705 ± 0.36 |
4.12 ± 0.634 | 8.365 ± 0.581 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.415 ± 0.381 | 3.038 ± 0.589 |
6.991 ± 1.037 | 3.246 ± 0.517 |
6.866 ± 0.516 | 8.198 ± 0.588 |
6.866 ± 0.769 | 7.532 ± 0.649 |
1.498 ± 0.393 | 3.454 ± 0.379 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
