
Acetobacter peroxydans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria;
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
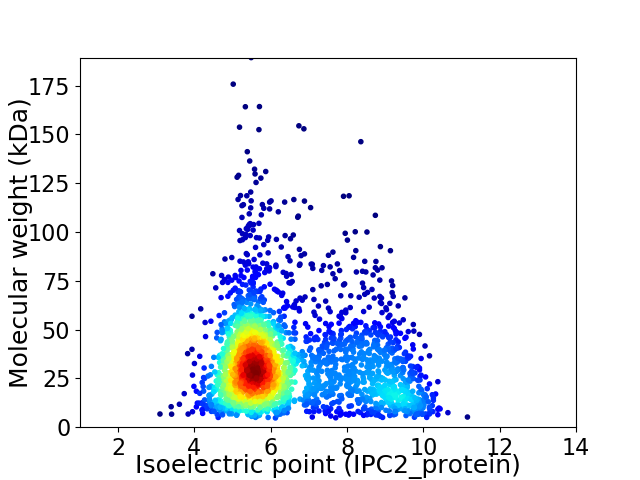
Virtual 2D-PAGE plot for 2457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y3TW46|A0A4Y3TW46_9PROT Transcriptional regulator OS=Acetobacter peroxydans OX=104098 GN=APE01nite_24130 PE=4 SV=1
MM1 pKa = 7.4SVFNALSTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.73KK37 pKa = 9.3ATTTAFQDD45 pKa = 4.29FVAGAGGNSASADD58 pKa = 3.19ISDD61 pKa = 3.73SVAAVTVQHH70 pKa = 6.61VDD72 pKa = 3.23NQGTSSTSTDD82 pKa = 3.16ALATSISGNGLFVVSKK98 pKa = 8.93GTTSGGQTYY107 pKa = 7.16YY108 pKa = 10.29TRR110 pKa = 11.84NGEE113 pKa = 4.17VYY115 pKa = 9.79EE116 pKa = 4.24NKK118 pKa = 10.34AGYY121 pKa = 9.43LVNTSGYY128 pKa = 8.12YY129 pKa = 10.29LEE131 pKa = 6.37GYY133 pKa = 8.6MVDD136 pKa = 4.0PSTGTLGTTLTQLNVSNVTFKK157 pKa = 9.98PTEE160 pKa = 4.22TTTLTSTSTVGTATGTNQTYY180 pKa = 7.15TTAPTTVYY188 pKa = 10.5DD189 pKa = 3.97SSGTAHH195 pKa = 6.89QIGLVWTQSTSDD207 pKa = 3.26TSKK210 pKa = 8.4WTVQAYY216 pKa = 9.86DD217 pKa = 3.69ADD219 pKa = 4.15GGGAISDD226 pKa = 4.52ASPSPYY232 pKa = 9.72IVTFDD237 pKa = 3.54SSTGAMTSVTDD248 pKa = 3.33ATGAVVANTTGTAAEE263 pKa = 4.13IPITATYY270 pKa = 10.49NGVAQTINLDD280 pKa = 3.56LGSIGSTSGTVVATAKK296 pKa = 10.51SGTPSTSQGTSLVVSGGSLAMDD318 pKa = 3.67SGGGAVNIGAQTGTNDD334 pKa = 3.39TYY336 pKa = 9.62TTTPLKK342 pKa = 10.72LSDD345 pKa = 3.65GSYY348 pKa = 11.21VSAVWTKK355 pKa = 11.14NPTTAANVNSEE366 pKa = 4.23QTWTVKK372 pKa = 10.66LVDD375 pKa = 4.81PYY377 pKa = 11.78SSATSDD383 pKa = 3.25STTAQVTFKK392 pKa = 11.14ADD394 pKa = 3.53GTIDD398 pKa = 3.95TINGSSTGSISFTDD412 pKa = 3.39SSGTTQTLNLTTSSVKK428 pKa = 10.53LGSSSPYY435 pKa = 9.5TDD437 pKa = 3.19TTAVTSDD444 pKa = 3.96SIATGTYY451 pKa = 8.67EE452 pKa = 4.02GAEE455 pKa = 4.1ITSDD459 pKa = 3.25GSVMAKK465 pKa = 10.45FSTGTQLIGKK475 pKa = 8.56VALANFANVNGLEE488 pKa = 4.24GVDD491 pKa = 3.54GQAYY495 pKa = 9.72VATTNSGNAQIGVSGQNGTGTLSVGYY521 pKa = 9.77VEE523 pKa = 6.33SSTTDD528 pKa = 3.22LTKK531 pKa = 10.76DD532 pKa = 3.4LSSLIVAQQAYY543 pKa = 5.96TANTKK548 pKa = 9.92VVTSANTMLQATIAMIQQ565 pKa = 2.99
MM1 pKa = 7.4SVFNALSTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.73KK37 pKa = 9.3ATTTAFQDD45 pKa = 4.29FVAGAGGNSASADD58 pKa = 3.19ISDD61 pKa = 3.73SVAAVTVQHH70 pKa = 6.61VDD72 pKa = 3.23NQGTSSTSTDD82 pKa = 3.16ALATSISGNGLFVVSKK98 pKa = 8.93GTTSGGQTYY107 pKa = 7.16YY108 pKa = 10.29TRR110 pKa = 11.84NGEE113 pKa = 4.17VYY115 pKa = 9.79EE116 pKa = 4.24NKK118 pKa = 10.34AGYY121 pKa = 9.43LVNTSGYY128 pKa = 8.12YY129 pKa = 10.29LEE131 pKa = 6.37GYY133 pKa = 8.6MVDD136 pKa = 4.0PSTGTLGTTLTQLNVSNVTFKK157 pKa = 9.98PTEE160 pKa = 4.22TTTLTSTSTVGTATGTNQTYY180 pKa = 7.15TTAPTTVYY188 pKa = 10.5DD189 pKa = 3.97SSGTAHH195 pKa = 6.89QIGLVWTQSTSDD207 pKa = 3.26TSKK210 pKa = 8.4WTVQAYY216 pKa = 9.86DD217 pKa = 3.69ADD219 pKa = 4.15GGGAISDD226 pKa = 4.52ASPSPYY232 pKa = 9.72IVTFDD237 pKa = 3.54SSTGAMTSVTDD248 pKa = 3.33ATGAVVANTTGTAAEE263 pKa = 4.13IPITATYY270 pKa = 10.49NGVAQTINLDD280 pKa = 3.56LGSIGSTSGTVVATAKK296 pKa = 10.51SGTPSTSQGTSLVVSGGSLAMDD318 pKa = 3.67SGGGAVNIGAQTGTNDD334 pKa = 3.39TYY336 pKa = 9.62TTTPLKK342 pKa = 10.72LSDD345 pKa = 3.65GSYY348 pKa = 11.21VSAVWTKK355 pKa = 11.14NPTTAANVNSEE366 pKa = 4.23QTWTVKK372 pKa = 10.66LVDD375 pKa = 4.81PYY377 pKa = 11.78SSATSDD383 pKa = 3.25STTAQVTFKK392 pKa = 11.14ADD394 pKa = 3.53GTIDD398 pKa = 3.95TINGSSTGSISFTDD412 pKa = 3.39SSGTTQTLNLTTSSVKK428 pKa = 10.53LGSSSPYY435 pKa = 9.5TDD437 pKa = 3.19TTAVTSDD444 pKa = 3.96SIATGTYY451 pKa = 8.67EE452 pKa = 4.02GAEE455 pKa = 4.1ITSDD459 pKa = 3.25GSVMAKK465 pKa = 10.45FSTGTQLIGKK475 pKa = 8.56VALANFANVNGLEE488 pKa = 4.24GVDD491 pKa = 3.54GQAYY495 pKa = 9.72VATTNSGNAQIGVSGQNGTGTLSVGYY521 pKa = 9.77VEE523 pKa = 6.33SSTTDD528 pKa = 3.22LTKK531 pKa = 10.76DD532 pKa = 3.4LSSLIVAQQAYY543 pKa = 5.96TANTKK548 pKa = 9.92VVTSANTMLQATIAMIQQ565 pKa = 2.99
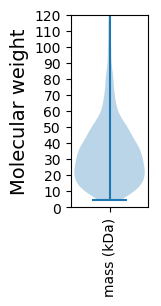
Molecular weight: 56.85 kDa
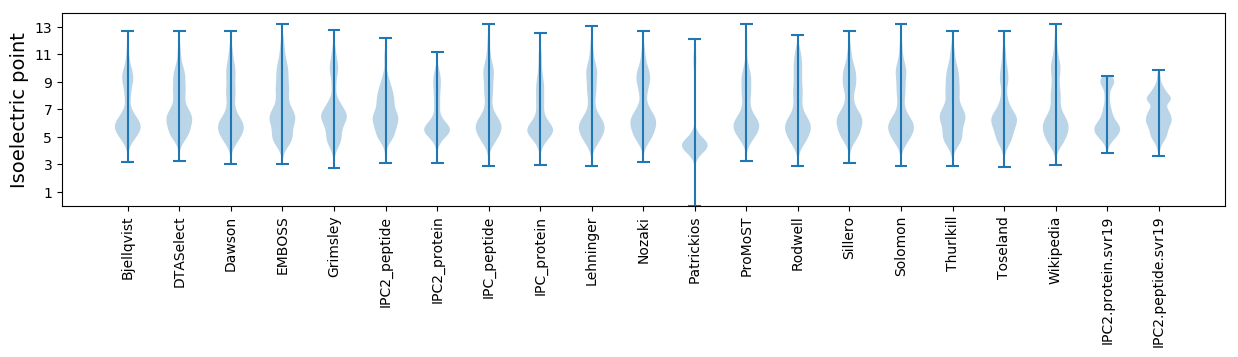
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y3TVP7|A0A4Y3TVP7_9PROT Uncharacterized protein OS=Acetobacter peroxydans OX=104098 GN=APE01nite_22830 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIANRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.46GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.32VIANRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.46GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
798204 |
43 |
1710 |
324.9 |
35.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.45 ± 0.072 | 1.096 ± 0.017 |
5.283 ± 0.034 | 5.349 ± 0.052 |
3.394 ± 0.029 | 8.37 ± 0.045 |
2.353 ± 0.022 | 4.539 ± 0.032 |
2.653 ± 0.042 | 10.635 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.021 | 2.534 ± 0.029 |
5.573 ± 0.038 | 3.692 ± 0.031 |
7.226 ± 0.046 | 5.597 ± 0.038 |
5.744 ± 0.039 | 7.404 ± 0.043 |
1.358 ± 0.02 | 2.152 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
