
Zetapapillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Zetapapillomavirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
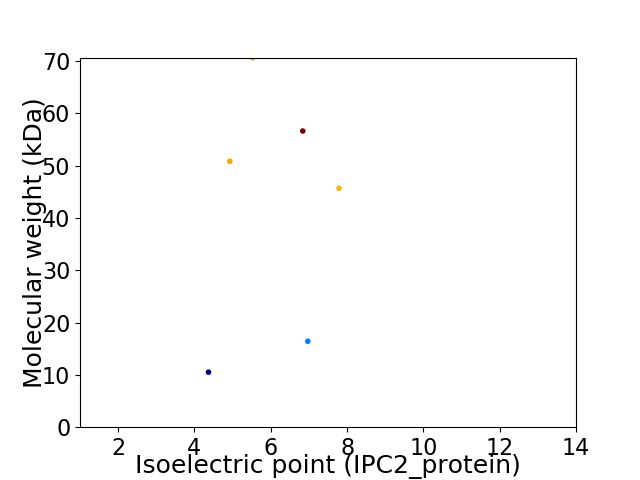
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8BE76|Q8BE76_9PAPI Protein E6 OS=Zetapapillomavirus 1 OX=333919 GN=E6 PE=3 SV=1
MM1 pKa = 7.66IGNGSPSLRR10 pKa = 11.84EE11 pKa = 3.71IVLSEE16 pKa = 4.4LPQSLADD23 pKa = 3.64PAEE26 pKa = 4.43AEE28 pKa = 4.24SEE30 pKa = 4.2EE31 pKa = 4.48EE32 pKa = 4.04EE33 pKa = 5.03VEE35 pKa = 4.29VEE37 pKa = 3.95LDD39 pKa = 3.34AVRR42 pKa = 11.84PQAPYY47 pKa = 10.4AVCTVCCRR55 pKa = 11.84CGEE58 pKa = 4.17KK59 pKa = 10.62VGLCVLATDD68 pKa = 4.21EE69 pKa = 5.31GIHH72 pKa = 6.0GLEE75 pKa = 3.84EE76 pKa = 4.25LLFEE80 pKa = 4.98ALQLFCAQCAPPIGRR95 pKa = 11.84HH96 pKa = 4.7GRR98 pKa = 3.45
MM1 pKa = 7.66IGNGSPSLRR10 pKa = 11.84EE11 pKa = 3.71IVLSEE16 pKa = 4.4LPQSLADD23 pKa = 3.64PAEE26 pKa = 4.43AEE28 pKa = 4.24SEE30 pKa = 4.2EE31 pKa = 4.48EE32 pKa = 4.04EE33 pKa = 5.03VEE35 pKa = 4.29VEE37 pKa = 3.95LDD39 pKa = 3.34AVRR42 pKa = 11.84PQAPYY47 pKa = 10.4AVCTVCCRR55 pKa = 11.84CGEE58 pKa = 4.17KK59 pKa = 10.62VGLCVLATDD68 pKa = 4.21EE69 pKa = 5.31GIHH72 pKa = 6.0GLEE75 pKa = 3.84EE76 pKa = 4.25LLFEE80 pKa = 4.98ALQLFCAQCAPPIGRR95 pKa = 11.84HH96 pKa = 4.7GRR98 pKa = 3.45
Molecular weight: 10.51 kDa
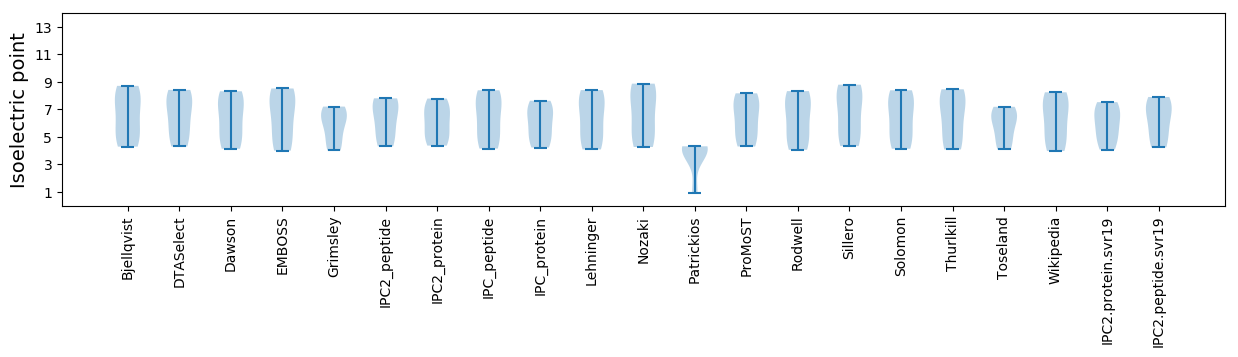
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8BE74|Q8BE74_9PAPI Replication protein E1 OS=Zetapapillomavirus 1 OX=333919 GN=E1 PE=3 SV=1
MM1 pKa = 7.42MEE3 pKa = 4.48TLRR6 pKa = 11.84QRR8 pKa = 11.84LDD10 pKa = 3.21AVQEE14 pKa = 4.0KK15 pKa = 10.52LMNLLEE21 pKa = 4.72EE22 pKa = 4.9GSSDD26 pKa = 4.07LSSQICYY33 pKa = 7.63WQAVRR38 pKa = 11.84KK39 pKa = 9.68EE40 pKa = 4.02NVLLYY45 pKa = 9.07YY46 pKa = 10.6ARR48 pKa = 11.84EE49 pKa = 4.09KK50 pKa = 10.94GLSRR54 pKa = 11.84LGLQMVPHH62 pKa = 6.81KK63 pKa = 10.68AVSQSQAKK71 pKa = 8.67QAIHH75 pKa = 6.38MEE77 pKa = 4.89LILLSLQGSSYY88 pKa = 9.39EE89 pKa = 4.03QEE91 pKa = 3.72PWTLSDD97 pKa = 4.25CSWEE101 pKa = 4.01RR102 pKa = 11.84WLQAPINCLKK112 pKa = 9.89KK113 pKa = 10.59DD114 pKa = 3.66PVIVEE119 pKa = 3.73VVYY122 pKa = 11.0DD123 pKa = 4.07GNSEE127 pKa = 3.9NANWYY132 pKa = 6.44TLWGLIYY139 pKa = 10.13YY140 pKa = 7.53QTFEE144 pKa = 4.39GDD146 pKa = 3.35WMCTRR151 pKa = 11.84GQCDD155 pKa = 3.0HH156 pKa = 6.96SGLYY160 pKa = 10.06YY161 pKa = 10.53EE162 pKa = 4.89EE163 pKa = 5.05EE164 pKa = 3.96GHH166 pKa = 5.99KK167 pKa = 9.97RR168 pKa = 11.84YY169 pKa = 9.47YY170 pKa = 10.48VHH172 pKa = 7.48FIDD175 pKa = 5.56DD176 pKa = 3.59AARR179 pKa = 11.84YY180 pKa = 9.13SKK182 pKa = 9.66TRR184 pKa = 11.84TWEE187 pKa = 3.86VRR189 pKa = 11.84CRR191 pKa = 11.84NQIYY195 pKa = 10.2LPSIPVTSTPPQSPSHH211 pKa = 6.77IDD213 pKa = 3.6LPDD216 pKa = 3.71GAAGGGPNQSPRR228 pKa = 11.84PGALAVSPQEE238 pKa = 4.14PPKK241 pKa = 10.55KK242 pKa = 9.74RR243 pKa = 11.84YY244 pKa = 9.44RR245 pKa = 11.84SPADD249 pKa = 3.41TVSSSRR255 pKa = 11.84LSGGLRR261 pKa = 11.84CPADD265 pKa = 2.85WCRR268 pKa = 11.84RR269 pKa = 11.84KK270 pKa = 9.89LQRR273 pKa = 11.84TSAPTWVPPSVSEE286 pKa = 4.32VPEE289 pKa = 4.16APEE292 pKa = 4.36GSVSEE297 pKa = 4.37TGGASPGVDD306 pKa = 2.73STTGRR311 pKa = 11.84GNDD314 pKa = 3.76PAPVPLEE321 pKa = 3.76AAFAPIVIFQGGTNQCKK338 pKa = 9.99CYY340 pKa = 9.91RR341 pKa = 11.84WRR343 pKa = 11.84LKK345 pKa = 10.08KK346 pKa = 9.96RR347 pKa = 11.84HH348 pKa = 6.22RR349 pKa = 11.84SLFVAITTTYY359 pKa = 10.23FWTGDD364 pKa = 3.1KK365 pKa = 10.92GGQRR369 pKa = 11.84VGNARR374 pKa = 11.84LMVTFSSDD382 pKa = 2.98LQRR385 pKa = 11.84RR386 pKa = 11.84LLLATVPPPRR396 pKa = 11.84GVTATSFTLTPSS408 pKa = 3.08
MM1 pKa = 7.42MEE3 pKa = 4.48TLRR6 pKa = 11.84QRR8 pKa = 11.84LDD10 pKa = 3.21AVQEE14 pKa = 4.0KK15 pKa = 10.52LMNLLEE21 pKa = 4.72EE22 pKa = 4.9GSSDD26 pKa = 4.07LSSQICYY33 pKa = 7.63WQAVRR38 pKa = 11.84KK39 pKa = 9.68EE40 pKa = 4.02NVLLYY45 pKa = 9.07YY46 pKa = 10.6ARR48 pKa = 11.84EE49 pKa = 4.09KK50 pKa = 10.94GLSRR54 pKa = 11.84LGLQMVPHH62 pKa = 6.81KK63 pKa = 10.68AVSQSQAKK71 pKa = 8.67QAIHH75 pKa = 6.38MEE77 pKa = 4.89LILLSLQGSSYY88 pKa = 9.39EE89 pKa = 4.03QEE91 pKa = 3.72PWTLSDD97 pKa = 4.25CSWEE101 pKa = 4.01RR102 pKa = 11.84WLQAPINCLKK112 pKa = 9.89KK113 pKa = 10.59DD114 pKa = 3.66PVIVEE119 pKa = 3.73VVYY122 pKa = 11.0DD123 pKa = 4.07GNSEE127 pKa = 3.9NANWYY132 pKa = 6.44TLWGLIYY139 pKa = 10.13YY140 pKa = 7.53QTFEE144 pKa = 4.39GDD146 pKa = 3.35WMCTRR151 pKa = 11.84GQCDD155 pKa = 3.0HH156 pKa = 6.96SGLYY160 pKa = 10.06YY161 pKa = 10.53EE162 pKa = 4.89EE163 pKa = 5.05EE164 pKa = 3.96GHH166 pKa = 5.99KK167 pKa = 9.97RR168 pKa = 11.84YY169 pKa = 9.47YY170 pKa = 10.48VHH172 pKa = 7.48FIDD175 pKa = 5.56DD176 pKa = 3.59AARR179 pKa = 11.84YY180 pKa = 9.13SKK182 pKa = 9.66TRR184 pKa = 11.84TWEE187 pKa = 3.86VRR189 pKa = 11.84CRR191 pKa = 11.84NQIYY195 pKa = 10.2LPSIPVTSTPPQSPSHH211 pKa = 6.77IDD213 pKa = 3.6LPDD216 pKa = 3.71GAAGGGPNQSPRR228 pKa = 11.84PGALAVSPQEE238 pKa = 4.14PPKK241 pKa = 10.55KK242 pKa = 9.74RR243 pKa = 11.84YY244 pKa = 9.44RR245 pKa = 11.84SPADD249 pKa = 3.41TVSSSRR255 pKa = 11.84LSGGLRR261 pKa = 11.84CPADD265 pKa = 2.85WCRR268 pKa = 11.84RR269 pKa = 11.84KK270 pKa = 9.89LQRR273 pKa = 11.84TSAPTWVPPSVSEE286 pKa = 4.32VPEE289 pKa = 4.16APEE292 pKa = 4.36GSVSEE297 pKa = 4.37TGGASPGVDD306 pKa = 2.73STTGRR311 pKa = 11.84GNDD314 pKa = 3.76PAPVPLEE321 pKa = 3.76AAFAPIVIFQGGTNQCKK338 pKa = 9.99CYY340 pKa = 9.91RR341 pKa = 11.84WRR343 pKa = 11.84LKK345 pKa = 10.08KK346 pKa = 9.96RR347 pKa = 11.84HH348 pKa = 6.22RR349 pKa = 11.84SLFVAITTTYY359 pKa = 10.23FWTGDD364 pKa = 3.1KK365 pKa = 10.92GGQRR369 pKa = 11.84VGNARR374 pKa = 11.84LMVTFSSDD382 pKa = 2.98LQRR385 pKa = 11.84RR386 pKa = 11.84LLLATVPPPRR396 pKa = 11.84GVTATSFTLTPSS408 pKa = 3.08
Molecular weight: 45.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2248 |
98 |
620 |
374.7 |
41.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.45 ± 0.488 | 2.224 ± 0.704 |
5.694 ± 0.568 | 6.361 ± 0.636 |
3.826 ± 0.572 | 7.829 ± 1.116 |
2.002 ± 0.113 | 3.692 ± 0.273 |
3.915 ± 0.734 | 8.763 ± 0.464 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.957 ± 0.226 | 3.781 ± 0.684 |
7.073 ± 0.708 | 4.181 ± 0.593 |
6.717 ± 0.362 | 7.473 ± 0.724 |
5.738 ± 0.53 | 7.251 ± 0.475 |
1.957 ± 0.358 | 3.114 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
