
Influenza A virus (A/duck/Jiangxi/5460/2014(mixed))
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Alphainfluenzavirus; Influenza A virus; mixed subtypes
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
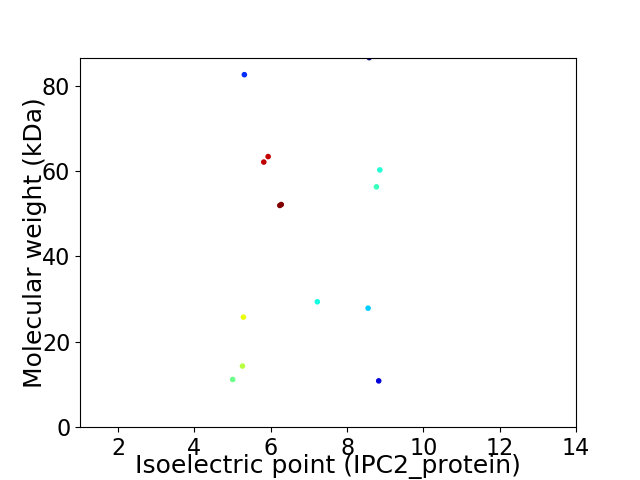
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UVB0|A0A0B4UVB0_9INFA Isoform of A0A0B4UWC0 Neuraminidase OS=Influenza A virus (A/duck/Jiangxi/5460/2014(mixed)) OX=1587846 GN=NA PE=3 SV=1
MM1 pKa = 7.38SLLTEE6 pKa = 4.4VEE8 pKa = 4.38TPTRR12 pKa = 11.84NGWEE16 pKa = 4.5CKK18 pKa = 10.41CNDD21 pKa = 3.43SSDD24 pKa = 4.01PLVIAASIIGILHH37 pKa = 7.1LILWILDD44 pKa = 3.39RR45 pKa = 11.84LFFKK49 pKa = 10.78CIYY52 pKa = 9.95RR53 pKa = 11.84RR54 pKa = 11.84LKK56 pKa = 10.55YY57 pKa = 10.03GLKK60 pKa = 10.09RR61 pKa = 11.84GPSTEE66 pKa = 4.02GVPEE70 pKa = 4.24SMRR73 pKa = 11.84EE74 pKa = 3.92EE75 pKa = 4.1YY76 pKa = 10.41RR77 pKa = 11.84QEE79 pKa = 3.74QQSAVDD85 pKa = 3.3VDD87 pKa = 4.47DD88 pKa = 3.92GHH90 pKa = 6.42FVNIEE95 pKa = 3.83LEE97 pKa = 4.08
MM1 pKa = 7.38SLLTEE6 pKa = 4.4VEE8 pKa = 4.38TPTRR12 pKa = 11.84NGWEE16 pKa = 4.5CKK18 pKa = 10.41CNDD21 pKa = 3.43SSDD24 pKa = 4.01PLVIAASIIGILHH37 pKa = 7.1LILWILDD44 pKa = 3.39RR45 pKa = 11.84LFFKK49 pKa = 10.78CIYY52 pKa = 9.95RR53 pKa = 11.84RR54 pKa = 11.84LKK56 pKa = 10.55YY57 pKa = 10.03GLKK60 pKa = 10.09RR61 pKa = 11.84GPSTEE66 pKa = 4.02GVPEE70 pKa = 4.24SMRR73 pKa = 11.84EE74 pKa = 3.92EE75 pKa = 4.1YY76 pKa = 10.41RR77 pKa = 11.84QEE79 pKa = 3.74QQSAVDD85 pKa = 3.3VDD87 pKa = 4.47DD88 pKa = 3.92GHH90 pKa = 6.42FVNIEE95 pKa = 3.83LEE97 pKa = 4.08
Molecular weight: 11.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UW38|A0A0B4UW38_9INFA Matrix protein 1 OS=Influenza A virus (A/duck/Jiangxi/5460/2014(mixed)) OX=1587846 GN=M1 PE=3 SV=1
MM1 pKa = 7.36EE2 pKa = 6.17RR3 pKa = 11.84IKK5 pKa = 10.51EE6 pKa = 4.0LRR8 pKa = 11.84DD9 pKa = 3.29LMSQSRR15 pKa = 11.84TRR17 pKa = 11.84EE18 pKa = 3.86ILTKK22 pKa = 8.81TTVDD26 pKa = 2.86HH27 pKa = 5.76MAIIKK32 pKa = 10.06KK33 pKa = 7.84YY34 pKa = 8.29TSGRR38 pKa = 11.84QEE40 pKa = 3.99KK41 pKa = 10.64NPALRR46 pKa = 11.84MKK48 pKa = 10.19WMMAMKK54 pKa = 10.45YY55 pKa = 10.07PITADD60 pKa = 3.19KK61 pKa = 10.67RR62 pKa = 11.84IMEE65 pKa = 4.94MIPEE69 pKa = 4.3RR70 pKa = 11.84NEE72 pKa = 3.52QGQTLWSKK80 pKa = 9.91TNDD83 pKa = 3.44AGSDD87 pKa = 3.19RR88 pKa = 11.84VMVSPLAVTWWNRR101 pKa = 11.84NGPTTSTVHH110 pKa = 5.17YY111 pKa = 8.49PKK113 pKa = 10.47VYY115 pKa = 8.68KK116 pKa = 10.24TYY118 pKa = 10.19FEE120 pKa = 3.9KK121 pKa = 11.0AEE123 pKa = 4.08RR124 pKa = 11.84LKK126 pKa = 10.93HH127 pKa = 4.77GTFGPVHH134 pKa = 6.7FRR136 pKa = 11.84NQVKK140 pKa = 9.47IRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84VDD146 pKa = 3.11INPGHH151 pKa = 7.33ADD153 pKa = 3.84LSAKK157 pKa = 9.4EE158 pKa = 3.98AQDD161 pKa = 3.52VIMEE165 pKa = 4.23VVFPNEE171 pKa = 3.21VGARR175 pKa = 11.84ILTSEE180 pKa = 4.22SQLTITKK187 pKa = 9.33EE188 pKa = 4.15KK189 pKa = 10.77KK190 pKa = 10.27EE191 pKa = 3.94EE192 pKa = 3.95LQDD195 pKa = 3.83CKK197 pKa = 10.5IAPLMVAYY205 pKa = 9.47MLEE208 pKa = 4.19RR209 pKa = 11.84EE210 pKa = 4.59LVRR213 pKa = 11.84KK214 pKa = 6.95XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXVLFQNWGVEE452 pKa = 4.28PIDD455 pKa = 3.95NVMGMIGILPDD466 pKa = 3.56MTPSTEE472 pKa = 3.52MSLRR476 pKa = 11.84GVRR479 pKa = 11.84VSKK482 pKa = 10.41MGVDD486 pKa = 4.11EE487 pKa = 4.4YY488 pKa = 11.53SSTEE492 pKa = 3.81RR493 pKa = 11.84VVVNIDD499 pKa = 2.92RR500 pKa = 11.84FLRR503 pKa = 11.84VRR505 pKa = 11.84DD506 pKa = 3.41QRR508 pKa = 11.84GNILLSPEE516 pKa = 4.37EE517 pKa = 3.98VSEE520 pKa = 4.32TQGTEE525 pKa = 3.64KK526 pKa = 10.4LTITYY531 pKa = 9.18SSSMMWEE538 pKa = 3.7INGPEE543 pKa = 3.98SVLVNTYY550 pKa = 7.57QWIIRR555 pKa = 11.84NWEE558 pKa = 3.94TVKK561 pKa = 10.56IQWSQDD567 pKa = 2.67PTMLYY572 pKa = 10.81NKK574 pKa = 9.63MEE576 pKa = 4.24FEE578 pKa = 4.66PFQSLVPKK586 pKa = 10.24AARR589 pKa = 11.84GQYY592 pKa = 10.02SGFVRR597 pKa = 11.84TLFQQMRR604 pKa = 11.84DD605 pKa = 3.48VLGTFDD611 pKa = 3.43TVQIIKK617 pKa = 10.36LLPFAAAPPEE627 pKa = 3.92QSRR630 pKa = 11.84MQFSSLTVNVRR641 pKa = 11.84GSGMRR646 pKa = 11.84ILVRR650 pKa = 11.84GNSPVFNYY658 pKa = 10.56NKK660 pKa = 8.71ATKK663 pKa = 10.14RR664 pKa = 11.84LTVLGKK670 pKa = 10.13DD671 pKa = 3.2AGALTEE677 pKa = 5.36DD678 pKa = 4.62PDD680 pKa = 3.67EE681 pKa = 4.45GTXGVEE687 pKa = 4.06SAVLRR692 pKa = 11.84GFLILGKK699 pKa = 9.47EE700 pKa = 4.26DD701 pKa = 3.46KK702 pKa = 10.5RR703 pKa = 11.84YY704 pKa = 10.65GPALSINEE712 pKa = 4.34LSNLAKK718 pKa = 10.63GEE720 pKa = 4.09KK721 pKa = 10.32ANVLIGQGDD730 pKa = 3.9VVLVMKK736 pKa = 10.39RR737 pKa = 11.84KK738 pKa = 9.29RR739 pKa = 11.84DD740 pKa = 3.62SSILTDD746 pKa = 3.31SQTATKK752 pKa = 9.73RR753 pKa = 11.84IRR755 pKa = 11.84MAINN759 pKa = 3.15
MM1 pKa = 7.36EE2 pKa = 6.17RR3 pKa = 11.84IKK5 pKa = 10.51EE6 pKa = 4.0LRR8 pKa = 11.84DD9 pKa = 3.29LMSQSRR15 pKa = 11.84TRR17 pKa = 11.84EE18 pKa = 3.86ILTKK22 pKa = 8.81TTVDD26 pKa = 2.86HH27 pKa = 5.76MAIIKK32 pKa = 10.06KK33 pKa = 7.84YY34 pKa = 8.29TSGRR38 pKa = 11.84QEE40 pKa = 3.99KK41 pKa = 10.64NPALRR46 pKa = 11.84MKK48 pKa = 10.19WMMAMKK54 pKa = 10.45YY55 pKa = 10.07PITADD60 pKa = 3.19KK61 pKa = 10.67RR62 pKa = 11.84IMEE65 pKa = 4.94MIPEE69 pKa = 4.3RR70 pKa = 11.84NEE72 pKa = 3.52QGQTLWSKK80 pKa = 9.91TNDD83 pKa = 3.44AGSDD87 pKa = 3.19RR88 pKa = 11.84VMVSPLAVTWWNRR101 pKa = 11.84NGPTTSTVHH110 pKa = 5.17YY111 pKa = 8.49PKK113 pKa = 10.47VYY115 pKa = 8.68KK116 pKa = 10.24TYY118 pKa = 10.19FEE120 pKa = 3.9KK121 pKa = 11.0AEE123 pKa = 4.08RR124 pKa = 11.84LKK126 pKa = 10.93HH127 pKa = 4.77GTFGPVHH134 pKa = 6.7FRR136 pKa = 11.84NQVKK140 pKa = 9.47IRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84VDD146 pKa = 3.11INPGHH151 pKa = 7.33ADD153 pKa = 3.84LSAKK157 pKa = 9.4EE158 pKa = 3.98AQDD161 pKa = 3.52VIMEE165 pKa = 4.23VVFPNEE171 pKa = 3.21VGARR175 pKa = 11.84ILTSEE180 pKa = 4.22SQLTITKK187 pKa = 9.33EE188 pKa = 4.15KK189 pKa = 10.77KK190 pKa = 10.27EE191 pKa = 3.94EE192 pKa = 3.95LQDD195 pKa = 3.83CKK197 pKa = 10.5IAPLMVAYY205 pKa = 9.47MLEE208 pKa = 4.19RR209 pKa = 11.84EE210 pKa = 4.59LVRR213 pKa = 11.84KK214 pKa = 6.95XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXVLFQNWGVEE452 pKa = 4.28PIDD455 pKa = 3.95NVMGMIGILPDD466 pKa = 3.56MTPSTEE472 pKa = 3.52MSLRR476 pKa = 11.84GVRR479 pKa = 11.84VSKK482 pKa = 10.41MGVDD486 pKa = 4.11EE487 pKa = 4.4YY488 pKa = 11.53SSTEE492 pKa = 3.81RR493 pKa = 11.84VVVNIDD499 pKa = 2.92RR500 pKa = 11.84FLRR503 pKa = 11.84VRR505 pKa = 11.84DD506 pKa = 3.41QRR508 pKa = 11.84GNILLSPEE516 pKa = 4.37EE517 pKa = 3.98VSEE520 pKa = 4.32TQGTEE525 pKa = 3.64KK526 pKa = 10.4LTITYY531 pKa = 9.18SSSMMWEE538 pKa = 3.7INGPEE543 pKa = 3.98SVLVNTYY550 pKa = 7.57QWIIRR555 pKa = 11.84NWEE558 pKa = 3.94TVKK561 pKa = 10.56IQWSQDD567 pKa = 2.67PTMLYY572 pKa = 10.81NKK574 pKa = 9.63MEE576 pKa = 4.24FEE578 pKa = 4.66PFQSLVPKK586 pKa = 10.24AARR589 pKa = 11.84GQYY592 pKa = 10.02SGFVRR597 pKa = 11.84TLFQQMRR604 pKa = 11.84DD605 pKa = 3.48VLGTFDD611 pKa = 3.43TVQIIKK617 pKa = 10.36LLPFAAAPPEE627 pKa = 3.92QSRR630 pKa = 11.84MQFSSLTVNVRR641 pKa = 11.84GSGMRR646 pKa = 11.84ILVRR650 pKa = 11.84GNSPVFNYY658 pKa = 10.56NKK660 pKa = 8.71ATKK663 pKa = 10.14RR664 pKa = 11.84LTVLGKK670 pKa = 10.13DD671 pKa = 3.2AGALTEE677 pKa = 5.36DD678 pKa = 4.62PDD680 pKa = 3.67EE681 pKa = 4.45GTXGVEE687 pKa = 4.06SAVLRR692 pKa = 11.84GFLILGKK699 pKa = 9.47EE700 pKa = 4.26DD701 pKa = 3.46KK702 pKa = 10.5RR703 pKa = 11.84YY704 pKa = 10.65GPALSINEE712 pKa = 4.34LSNLAKK718 pKa = 10.63GEE720 pKa = 4.09KK721 pKa = 10.32ANVLIGQGDD730 pKa = 3.9VVLVMKK736 pKa = 10.39RR737 pKa = 11.84KK738 pKa = 9.29RR739 pKa = 11.84DD740 pKa = 3.62SSILTDD746 pKa = 3.31SQTATKK752 pKa = 9.73RR753 pKa = 11.84IRR755 pKa = 11.84MAINN759 pKa = 3.15
Molecular weight: 60.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5838 |
90 |
759 |
417.0 |
45.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.379 ± 0.417 | 2.073 ± 0.406 |
4.693 ± 0.297 | 6.903 ± 0.641 |
3.888 ± 0.306 | 6.629 ± 0.573 |
1.627 ± 0.172 | 6.492 ± 0.475 |
5.464 ± 0.381 | 7.211 ± 0.494 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.374 ± 0.399 | 5.344 ± 0.55 |
3.803 ± 0.251 | 3.905 ± 0.368 |
6.355 ± 0.525 | 6.869 ± 0.319 |
6.475 ± 0.384 | 5.396 ± 0.399 |
1.679 ± 0.178 | 2.467 ± 0.209 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
