
Mycobacterium talmoniae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
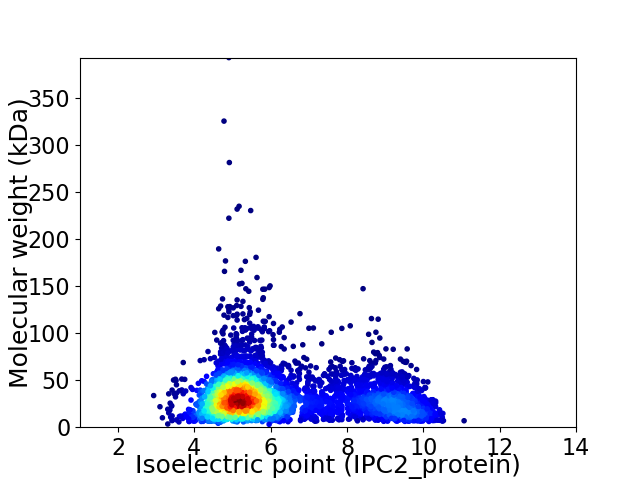
Virtual 2D-PAGE plot for 5059 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S1NLT2|A0A1S1NLT2_9MYCO Uncharacterized protein OS=Mycobacterium talmoniae OX=1858794 GN=BKN37_07030 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84NQLARR7 pKa = 11.84FRR9 pKa = 11.84GPTILLATAVAGVIAVAPVTPSVRR33 pKa = 11.84PGQLPDD39 pKa = 3.17IRR41 pKa = 11.84LTADD45 pKa = 3.43DD46 pKa = 5.48ASWQPALDD54 pKa = 3.71QLLDD58 pKa = 3.56AMQQAVAQDD67 pKa = 3.52SALPFATPDD76 pKa = 3.23SAATLMTSLQNTNYY90 pKa = 8.91EE91 pKa = 4.19LLMAGISRR99 pKa = 11.84VTEE102 pKa = 4.14LFNGWFFQSPEE113 pKa = 4.15VVGGDD118 pKa = 3.27AADD121 pKa = 3.61PRR123 pKa = 11.84QYY125 pKa = 11.06FQFFTPDD132 pKa = 2.61IYY134 pKa = 11.44YY135 pKa = 10.32HH136 pKa = 6.0ATAGLAPGATYY147 pKa = 10.82EE148 pKa = 4.13LTGTVGGGTEE158 pKa = 3.92ALAIATEE165 pKa = 4.69EE166 pKa = 4.26ITGSTAVSKK175 pKa = 11.11SSLEE179 pKa = 3.99LGHH182 pKa = 6.48NLVVNPNGTFTVYY195 pKa = 10.33IGPTDD200 pKa = 3.47PSGAVNYY207 pKa = 10.87LNDD210 pKa = 3.86TDD212 pKa = 4.3ATAQGDD218 pKa = 3.86ASLLIRR224 pKa = 11.84DD225 pKa = 3.93MLGNSAQGPTNVSIHH240 pKa = 6.28CVADD244 pKa = 3.91CPAFFAIPSTGLFPGAGDD262 pKa = 3.86SAADD266 pKa = 3.44AAGTSGTQITSLDD279 pKa = 3.79SLLTTLFTAFANIVGPFNSEE299 pKa = 3.58NMGLAQIAGIEE310 pKa = 4.2LPANTMGALTPEE322 pKa = 4.19TSLFATGLPSADD334 pKa = 2.94VSAGNFDD341 pKa = 5.16LDD343 pKa = 3.82PGQALIVKK351 pKa = 9.39VPDD354 pKa = 3.27VAAGYY359 pKa = 10.43SGIEE363 pKa = 3.92LMNVYY368 pKa = 10.17GAGLPSTLAQTTLNNTQAFHH388 pKa = 7.29DD389 pKa = 4.15ADD391 pKa = 4.14GYY393 pKa = 8.75TYY395 pKa = 11.07YY396 pKa = 10.82VVSATNPGVANWLDD410 pKa = 3.53SSGVTRR416 pKa = 11.84GEE418 pKa = 3.41IFARR422 pKa = 11.84FEE424 pKa = 4.02NLPDD428 pKa = 3.81GTDD431 pKa = 3.48PTGLPVTTQVVPVDD445 pKa = 4.1EE446 pKa = 4.57VSHH449 pKa = 5.78YY450 pKa = 11.07LPADD454 pKa = 3.7TPTVSPEE461 pKa = 3.46EE462 pKa = 4.17YY463 pKa = 10.17AADD466 pKa = 3.33MAQRR470 pKa = 11.84VFSYY474 pKa = 10.59DD475 pKa = 3.18YY476 pKa = 11.01ALDD479 pKa = 3.39VSRR482 pKa = 11.84EE483 pKa = 3.87HH484 pKa = 6.91AQPDD488 pKa = 3.62WVIQQLLLHH497 pKa = 6.59GLQGLMGTDD506 pKa = 2.99NYY508 pKa = 11.42DD509 pKa = 3.12AVFGSEE515 pKa = 4.12PVTPLALRR523 pKa = 11.84FSDD526 pKa = 4.34ALSPDD531 pKa = 4.06FGAVLHH537 pKa = 7.09DD538 pKa = 3.86VLTNPVGSLEE548 pKa = 4.16AVINNVPLLASDD560 pKa = 3.58IGLPMEE566 pKa = 4.43LAVGQTVLSLLLPQQLGSLLDD587 pKa = 3.96SVLLDD592 pKa = 3.86PNSGIVAGLLNARR605 pKa = 11.84DD606 pKa = 4.18DD607 pKa = 4.04LATAILTANGGFPTEE622 pKa = 4.87LGVQAAQEE630 pKa = 4.07WANMAEE636 pKa = 4.77LVSSTPITTLADD648 pKa = 3.8LGSLFSLVDD657 pKa = 4.03FPWW660 pKa = 5.08
MM1 pKa = 7.88RR2 pKa = 11.84NQLARR7 pKa = 11.84FRR9 pKa = 11.84GPTILLATAVAGVIAVAPVTPSVRR33 pKa = 11.84PGQLPDD39 pKa = 3.17IRR41 pKa = 11.84LTADD45 pKa = 3.43DD46 pKa = 5.48ASWQPALDD54 pKa = 3.71QLLDD58 pKa = 3.56AMQQAVAQDD67 pKa = 3.52SALPFATPDD76 pKa = 3.23SAATLMTSLQNTNYY90 pKa = 8.91EE91 pKa = 4.19LLMAGISRR99 pKa = 11.84VTEE102 pKa = 4.14LFNGWFFQSPEE113 pKa = 4.15VVGGDD118 pKa = 3.27AADD121 pKa = 3.61PRR123 pKa = 11.84QYY125 pKa = 11.06FQFFTPDD132 pKa = 2.61IYY134 pKa = 11.44YY135 pKa = 10.32HH136 pKa = 6.0ATAGLAPGATYY147 pKa = 10.82EE148 pKa = 4.13LTGTVGGGTEE158 pKa = 3.92ALAIATEE165 pKa = 4.69EE166 pKa = 4.26ITGSTAVSKK175 pKa = 11.11SSLEE179 pKa = 3.99LGHH182 pKa = 6.48NLVVNPNGTFTVYY195 pKa = 10.33IGPTDD200 pKa = 3.47PSGAVNYY207 pKa = 10.87LNDD210 pKa = 3.86TDD212 pKa = 4.3ATAQGDD218 pKa = 3.86ASLLIRR224 pKa = 11.84DD225 pKa = 3.93MLGNSAQGPTNVSIHH240 pKa = 6.28CVADD244 pKa = 3.91CPAFFAIPSTGLFPGAGDD262 pKa = 3.86SAADD266 pKa = 3.44AAGTSGTQITSLDD279 pKa = 3.79SLLTTLFTAFANIVGPFNSEE299 pKa = 3.58NMGLAQIAGIEE310 pKa = 4.2LPANTMGALTPEE322 pKa = 4.19TSLFATGLPSADD334 pKa = 2.94VSAGNFDD341 pKa = 5.16LDD343 pKa = 3.82PGQALIVKK351 pKa = 9.39VPDD354 pKa = 3.27VAAGYY359 pKa = 10.43SGIEE363 pKa = 3.92LMNVYY368 pKa = 10.17GAGLPSTLAQTTLNNTQAFHH388 pKa = 7.29DD389 pKa = 4.15ADD391 pKa = 4.14GYY393 pKa = 8.75TYY395 pKa = 11.07YY396 pKa = 10.82VVSATNPGVANWLDD410 pKa = 3.53SSGVTRR416 pKa = 11.84GEE418 pKa = 3.41IFARR422 pKa = 11.84FEE424 pKa = 4.02NLPDD428 pKa = 3.81GTDD431 pKa = 3.48PTGLPVTTQVVPVDD445 pKa = 4.1EE446 pKa = 4.57VSHH449 pKa = 5.78YY450 pKa = 11.07LPADD454 pKa = 3.7TPTVSPEE461 pKa = 3.46EE462 pKa = 4.17YY463 pKa = 10.17AADD466 pKa = 3.33MAQRR470 pKa = 11.84VFSYY474 pKa = 10.59DD475 pKa = 3.18YY476 pKa = 11.01ALDD479 pKa = 3.39VSRR482 pKa = 11.84EE483 pKa = 3.87HH484 pKa = 6.91AQPDD488 pKa = 3.62WVIQQLLLHH497 pKa = 6.59GLQGLMGTDD506 pKa = 2.99NYY508 pKa = 11.42DD509 pKa = 3.12AVFGSEE515 pKa = 4.12PVTPLALRR523 pKa = 11.84FSDD526 pKa = 4.34ALSPDD531 pKa = 4.06FGAVLHH537 pKa = 7.09DD538 pKa = 3.86VLTNPVGSLEE548 pKa = 4.16AVINNVPLLASDD560 pKa = 3.58IGLPMEE566 pKa = 4.43LAVGQTVLSLLLPQQLGSLLDD587 pKa = 3.96SVLLDD592 pKa = 3.86PNSGIVAGLLNARR605 pKa = 11.84DD606 pKa = 4.18DD607 pKa = 4.04LATAILTANGGFPTEE622 pKa = 4.87LGVQAAQEE630 pKa = 4.07WANMAEE636 pKa = 4.77LVSSTPITTLADD648 pKa = 3.8LGSLFSLVDD657 pKa = 4.03FPWW660 pKa = 5.08
Molecular weight: 68.82 kDa
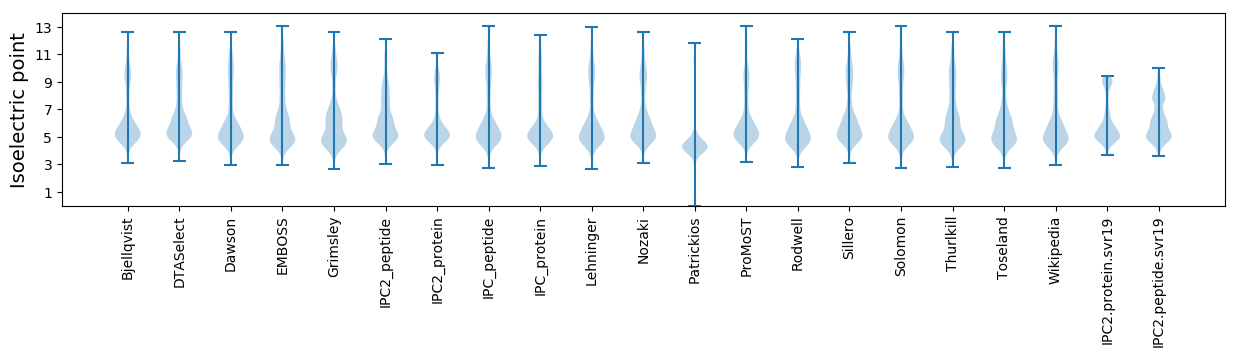
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S1NCA5|A0A1S1NCA5_9MYCO Transporter OS=Mycobacterium talmoniae OX=1858794 GN=BKN37_15780 PE=3 SV=1
MM1 pKa = 7.52AFVLAHH7 pKa = 7.09PIPTARR13 pKa = 11.84RR14 pKa = 11.84STLLHH19 pKa = 6.74AGRR22 pKa = 11.84TAASRR27 pKa = 11.84LWARR31 pKa = 11.84RR32 pKa = 11.84TPQQRR37 pKa = 11.84AAAYY41 pKa = 8.43VSRR44 pKa = 11.84MPVAVITASLGGHH57 pKa = 4.94PTHH60 pKa = 7.47RR61 pKa = 11.84RR62 pKa = 11.84GG63 pKa = 3.62
MM1 pKa = 7.52AFVLAHH7 pKa = 7.09PIPTARR13 pKa = 11.84RR14 pKa = 11.84STLLHH19 pKa = 6.74AGRR22 pKa = 11.84TAASRR27 pKa = 11.84LWARR31 pKa = 11.84RR32 pKa = 11.84TPQQRR37 pKa = 11.84AAAYY41 pKa = 8.43VSRR44 pKa = 11.84MPVAVITASLGGHH57 pKa = 4.94PTHH60 pKa = 7.47RR61 pKa = 11.84RR62 pKa = 11.84GG63 pKa = 3.62
Molecular weight: 6.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1588304 |
29 |
3683 |
314.0 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.627 ± 0.052 | 0.819 ± 0.01 |
6.365 ± 0.027 | 4.91 ± 0.03 |
3.017 ± 0.016 | 8.819 ± 0.032 |
2.308 ± 0.018 | 4.081 ± 0.022 |
2.077 ± 0.022 | 9.997 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.963 ± 0.014 | 2.15 ± 0.019 |
5.95 ± 0.029 | 3.129 ± 0.019 |
7.388 ± 0.035 | 4.95 ± 0.023 |
6.062 ± 0.024 | 8.658 ± 0.035 |
1.526 ± 0.015 | 2.205 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
