
Red clover powdery mildew-associated totivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
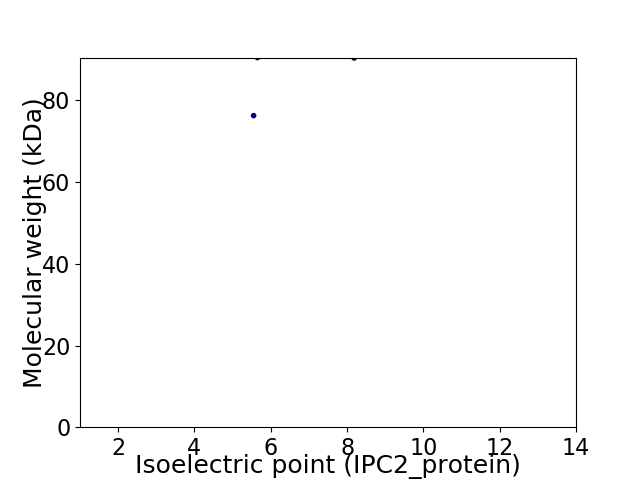
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
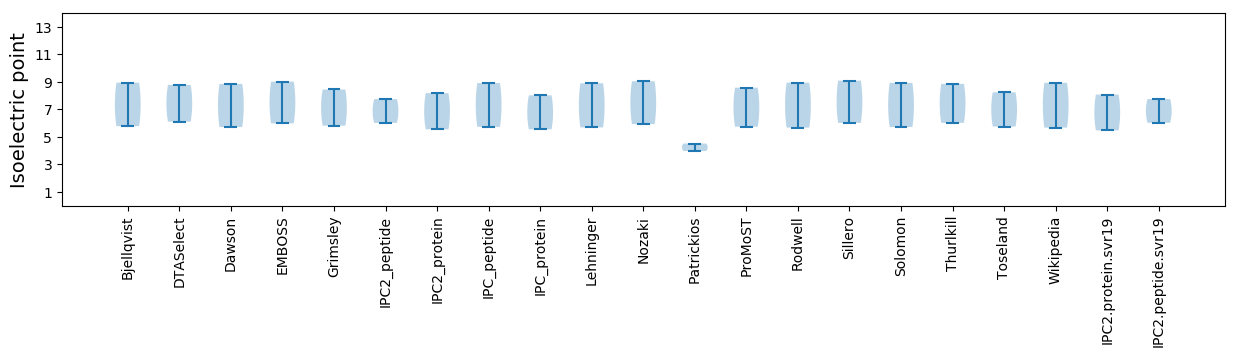
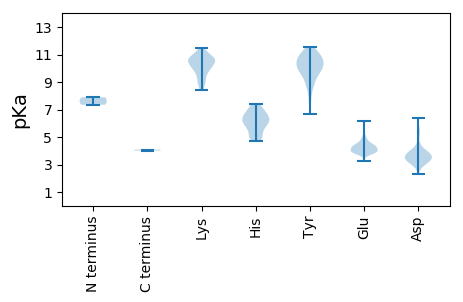
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3Q2B6|A0A0S3Q2B6_9VIRU Capsid protein OS=Red clover powdery mildew-associated totivirus 1 OX=1714362 GN=CP PE=4 SV=1
MM1 pKa = 7.89DD2 pKa = 6.35KK3 pKa = 10.37IIKK6 pKa = 9.92HH7 pKa = 4.71VFEE10 pKa = 4.59SEE12 pKa = 3.85GMAGMPIKK20 pKa = 10.68HH21 pKa = 6.3EE22 pKa = 4.42SGNYY26 pKa = 6.65NTLLKK31 pKa = 10.99GNLAVDD37 pKa = 3.59VNGDD41 pKa = 3.49KK42 pKa = 10.84NIKK45 pKa = 9.1KK46 pKa = 8.67LTMAQSMTYY55 pKa = 10.1LGRR58 pKa = 11.84STVVMGSGSSDD69 pKa = 3.34LEE71 pKa = 4.6GINKK75 pKa = 9.2FCLTPEE81 pKa = 4.26GNLNIVNVLATVAKK95 pKa = 9.97EE96 pKa = 3.91SGMPLNEE103 pKa = 3.76QHH105 pKa = 7.15HH106 pKa = 6.3KK107 pKa = 10.43LEE109 pKa = 4.17YY110 pKa = 8.84MKK112 pKa = 10.84TMSWADD118 pKa = 3.24NHH120 pKa = 6.71VSLLVNLMRR129 pKa = 11.84FAILLEE135 pKa = 4.23LSEE138 pKa = 4.49SSGGVGLKK146 pKa = 10.45GALPKK151 pKa = 11.16YY152 pKa = 9.51NDD154 pKa = 3.16GVVAVNRR161 pKa = 11.84TALLPEE167 pKa = 4.97EE168 pKa = 4.49YY169 pKa = 9.72PVGGDD174 pKa = 3.24MTWFTRR180 pKa = 11.84FSRR183 pKa = 11.84QDD185 pKa = 3.08MPDD188 pKa = 3.09VVRR191 pKa = 11.84VEE193 pKa = 4.89EE194 pKa = 4.38YY195 pKa = 10.34CAPQSDD201 pKa = 4.1VCLDD205 pKa = 3.6LRR207 pKa = 11.84SLTEE211 pKa = 3.91TEE213 pKa = 3.89AVIVLLCCGKK223 pKa = 9.65WYY225 pKa = 9.87RR226 pKa = 11.84QSRR229 pKa = 11.84FLLDD233 pKa = 3.85FDD235 pKa = 5.52CPILTSKK242 pKa = 10.74LYY244 pKa = 10.36YY245 pKa = 10.51RR246 pKa = 11.84FDD248 pKa = 3.28KK249 pKa = 11.08RR250 pKa = 11.84IDD252 pKa = 3.26IGNFDD257 pKa = 3.55QTSDD261 pKa = 3.82GRR263 pKa = 11.84VTQPKK268 pKa = 9.83LPKK271 pKa = 10.27AKK273 pKa = 9.97EE274 pKa = 3.83VWRR277 pKa = 11.84TMSRR281 pKa = 11.84YY282 pKa = 8.96LAANRR287 pKa = 11.84LYY289 pKa = 10.67GQYY292 pKa = 8.97STALCLVAQTMCQFIPATAEE312 pKa = 3.9GNVWLTQQRR321 pKa = 11.84VIHH324 pKa = 6.4LPALKK329 pKa = 9.82SVRR332 pKa = 11.84GALPFINEE340 pKa = 4.44DD341 pKa = 3.42EE342 pKa = 4.46PALVSQRR349 pKa = 11.84SLNEE353 pKa = 3.22WGAFSTDD360 pKa = 2.95VTKK363 pKa = 11.37YY364 pKa = 10.68NLLGNILVQAVQTGIAVRR382 pKa = 11.84ALRR385 pKa = 11.84FRR387 pKa = 11.84NEE389 pKa = 4.35DD390 pKa = 3.46NPEE393 pKa = 3.93DD394 pKa = 4.13LYY396 pKa = 10.63RR397 pKa = 11.84TYY399 pKa = 10.96DD400 pKa = 3.35TVTNPAVMYY409 pKa = 9.99GALCSEE415 pKa = 5.07ALRR418 pKa = 11.84CPVPLAGMSGVFVEE432 pKa = 6.18FEE434 pKa = 4.16LPDD437 pKa = 3.33EE438 pKa = 4.42RR439 pKa = 11.84EE440 pKa = 4.04EE441 pKa = 4.33VEE443 pKa = 5.07LEE445 pKa = 4.21FPVLVYY451 pKa = 10.5DD452 pKa = 4.61PKK454 pKa = 10.85RR455 pKa = 11.84CGSTIEE461 pKa = 3.92KK462 pKa = 10.51RR463 pKa = 11.84RR464 pKa = 11.84DD465 pKa = 3.22TEE467 pKa = 4.06YY468 pKa = 11.12LQLNSIPMAGVPTLLLPLNPIGKK491 pKa = 8.65MSPFNLRR498 pKa = 11.84GVFDD502 pKa = 3.65TADD505 pKa = 3.6LKK507 pKa = 9.1RR508 pKa = 11.84TKK510 pKa = 10.04MGWEE514 pKa = 3.86TDD516 pKa = 4.06LYY518 pKa = 10.99SAWSWAWLCRR528 pKa = 11.84LCGYY532 pKa = 9.55DD533 pKa = 2.77IQMRR537 pKa = 11.84QQYY540 pKa = 8.79GQSCGSQGDD549 pKa = 4.1YY550 pKa = 11.25SPDD553 pKa = 3.93ASWTWPILGSVGHH566 pKa = 6.34NGIVCISKK574 pKa = 10.32LVEE577 pKa = 4.35RR578 pKa = 11.84GNSFIDD584 pKa = 4.19LPPLHH589 pKa = 7.11SKK591 pKa = 9.84SCHH594 pKa = 4.99SVINYY599 pKa = 8.54KK600 pKa = 10.46FSINSHH606 pKa = 4.99SVSLPEE612 pKa = 3.81RR613 pKa = 11.84RR614 pKa = 11.84QPEE617 pKa = 4.74LIGDD621 pKa = 4.47RR622 pKa = 11.84YY623 pKa = 10.56SAAPVLNLRR632 pKa = 11.84HH633 pKa = 6.15INIATTTEE641 pKa = 3.92IIKK644 pKa = 10.76LKK646 pKa = 10.81GFIEE650 pKa = 4.45RR651 pKa = 11.84SEE653 pKa = 4.26SDD655 pKa = 3.48FRR657 pKa = 11.84FADD660 pKa = 3.98DD661 pKa = 3.69VQAGQIPPPEE671 pKa = 4.34VPHH674 pKa = 5.95VASDD678 pKa = 3.72VIEE681 pKa = 4.09
MM1 pKa = 7.89DD2 pKa = 6.35KK3 pKa = 10.37IIKK6 pKa = 9.92HH7 pKa = 4.71VFEE10 pKa = 4.59SEE12 pKa = 3.85GMAGMPIKK20 pKa = 10.68HH21 pKa = 6.3EE22 pKa = 4.42SGNYY26 pKa = 6.65NTLLKK31 pKa = 10.99GNLAVDD37 pKa = 3.59VNGDD41 pKa = 3.49KK42 pKa = 10.84NIKK45 pKa = 9.1KK46 pKa = 8.67LTMAQSMTYY55 pKa = 10.1LGRR58 pKa = 11.84STVVMGSGSSDD69 pKa = 3.34LEE71 pKa = 4.6GINKK75 pKa = 9.2FCLTPEE81 pKa = 4.26GNLNIVNVLATVAKK95 pKa = 9.97EE96 pKa = 3.91SGMPLNEE103 pKa = 3.76QHH105 pKa = 7.15HH106 pKa = 6.3KK107 pKa = 10.43LEE109 pKa = 4.17YY110 pKa = 8.84MKK112 pKa = 10.84TMSWADD118 pKa = 3.24NHH120 pKa = 6.71VSLLVNLMRR129 pKa = 11.84FAILLEE135 pKa = 4.23LSEE138 pKa = 4.49SSGGVGLKK146 pKa = 10.45GALPKK151 pKa = 11.16YY152 pKa = 9.51NDD154 pKa = 3.16GVVAVNRR161 pKa = 11.84TALLPEE167 pKa = 4.97EE168 pKa = 4.49YY169 pKa = 9.72PVGGDD174 pKa = 3.24MTWFTRR180 pKa = 11.84FSRR183 pKa = 11.84QDD185 pKa = 3.08MPDD188 pKa = 3.09VVRR191 pKa = 11.84VEE193 pKa = 4.89EE194 pKa = 4.38YY195 pKa = 10.34CAPQSDD201 pKa = 4.1VCLDD205 pKa = 3.6LRR207 pKa = 11.84SLTEE211 pKa = 3.91TEE213 pKa = 3.89AVIVLLCCGKK223 pKa = 9.65WYY225 pKa = 9.87RR226 pKa = 11.84QSRR229 pKa = 11.84FLLDD233 pKa = 3.85FDD235 pKa = 5.52CPILTSKK242 pKa = 10.74LYY244 pKa = 10.36YY245 pKa = 10.51RR246 pKa = 11.84FDD248 pKa = 3.28KK249 pKa = 11.08RR250 pKa = 11.84IDD252 pKa = 3.26IGNFDD257 pKa = 3.55QTSDD261 pKa = 3.82GRR263 pKa = 11.84VTQPKK268 pKa = 9.83LPKK271 pKa = 10.27AKK273 pKa = 9.97EE274 pKa = 3.83VWRR277 pKa = 11.84TMSRR281 pKa = 11.84YY282 pKa = 8.96LAANRR287 pKa = 11.84LYY289 pKa = 10.67GQYY292 pKa = 8.97STALCLVAQTMCQFIPATAEE312 pKa = 3.9GNVWLTQQRR321 pKa = 11.84VIHH324 pKa = 6.4LPALKK329 pKa = 9.82SVRR332 pKa = 11.84GALPFINEE340 pKa = 4.44DD341 pKa = 3.42EE342 pKa = 4.46PALVSQRR349 pKa = 11.84SLNEE353 pKa = 3.22WGAFSTDD360 pKa = 2.95VTKK363 pKa = 11.37YY364 pKa = 10.68NLLGNILVQAVQTGIAVRR382 pKa = 11.84ALRR385 pKa = 11.84FRR387 pKa = 11.84NEE389 pKa = 4.35DD390 pKa = 3.46NPEE393 pKa = 3.93DD394 pKa = 4.13LYY396 pKa = 10.63RR397 pKa = 11.84TYY399 pKa = 10.96DD400 pKa = 3.35TVTNPAVMYY409 pKa = 9.99GALCSEE415 pKa = 5.07ALRR418 pKa = 11.84CPVPLAGMSGVFVEE432 pKa = 6.18FEE434 pKa = 4.16LPDD437 pKa = 3.33EE438 pKa = 4.42RR439 pKa = 11.84EE440 pKa = 4.04EE441 pKa = 4.33VEE443 pKa = 5.07LEE445 pKa = 4.21FPVLVYY451 pKa = 10.5DD452 pKa = 4.61PKK454 pKa = 10.85RR455 pKa = 11.84CGSTIEE461 pKa = 3.92KK462 pKa = 10.51RR463 pKa = 11.84RR464 pKa = 11.84DD465 pKa = 3.22TEE467 pKa = 4.06YY468 pKa = 11.12LQLNSIPMAGVPTLLLPLNPIGKK491 pKa = 8.65MSPFNLRR498 pKa = 11.84GVFDD502 pKa = 3.65TADD505 pKa = 3.6LKK507 pKa = 9.1RR508 pKa = 11.84TKK510 pKa = 10.04MGWEE514 pKa = 3.86TDD516 pKa = 4.06LYY518 pKa = 10.99SAWSWAWLCRR528 pKa = 11.84LCGYY532 pKa = 9.55DD533 pKa = 2.77IQMRR537 pKa = 11.84QQYY540 pKa = 8.79GQSCGSQGDD549 pKa = 4.1YY550 pKa = 11.25SPDD553 pKa = 3.93ASWTWPILGSVGHH566 pKa = 6.34NGIVCISKK574 pKa = 10.32LVEE577 pKa = 4.35RR578 pKa = 11.84GNSFIDD584 pKa = 4.19LPPLHH589 pKa = 7.11SKK591 pKa = 9.84SCHH594 pKa = 4.99SVINYY599 pKa = 8.54KK600 pKa = 10.46FSINSHH606 pKa = 4.99SVSLPEE612 pKa = 3.81RR613 pKa = 11.84RR614 pKa = 11.84QPEE617 pKa = 4.74LIGDD621 pKa = 4.47RR622 pKa = 11.84YY623 pKa = 10.56SAAPVLNLRR632 pKa = 11.84HH633 pKa = 6.15INIATTTEE641 pKa = 3.92IIKK644 pKa = 10.76LKK646 pKa = 10.81GFIEE650 pKa = 4.45RR651 pKa = 11.84SEE653 pKa = 4.26SDD655 pKa = 3.48FRR657 pKa = 11.84FADD660 pKa = 3.98DD661 pKa = 3.69VQAGQIPPPEE671 pKa = 4.34VPHH674 pKa = 5.95VASDD678 pKa = 3.72VIEE681 pKa = 4.09
Molecular weight: 76.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3Q2B6|A0A0S3Q2B6_9VIRU Capsid protein OS=Red clover powdery mildew-associated totivirus 1 OX=1714362 GN=CP PE=4 SV=1
MM1 pKa = 7.35WPVFTRR7 pKa = 11.84MSEE10 pKa = 3.84NRR12 pKa = 11.84LEE14 pKa = 4.06VVDD17 pKa = 3.76RR18 pKa = 11.84HH19 pKa = 5.93KK20 pKa = 10.51CTHH23 pKa = 5.51VLVGVEE29 pKa = 4.44EE30 pKa = 5.03DD31 pKa = 3.53TMTWQGNDD39 pKa = 2.33IMYY42 pKa = 10.43AGQEE46 pKa = 4.29CVRR49 pKa = 11.84TCSYY53 pKa = 11.55GDD55 pKa = 3.31GGTRR59 pKa = 11.84AHH61 pKa = 6.44YY62 pKa = 10.53IKK64 pKa = 10.18MNKK67 pKa = 8.66PVSLLPPIVKK77 pKa = 9.36QRR79 pKa = 11.84LSAAYY84 pKa = 10.12SMVGGYY90 pKa = 10.38VYY92 pKa = 11.01GDD94 pKa = 3.54KK95 pKa = 11.27DD96 pKa = 3.29EE97 pKa = 4.67LASVRR102 pKa = 11.84QILKK106 pKa = 9.66PLKK109 pKa = 9.55QNYY112 pKa = 8.55KK113 pKa = 9.77HH114 pKa = 6.65DD115 pKa = 3.94VKK117 pKa = 11.0FSIRR121 pKa = 11.84SLTYY125 pKa = 9.96KK126 pKa = 10.53KK127 pKa = 10.5SATTGEE133 pKa = 4.3HH134 pKa = 5.44HH135 pKa = 5.79THH137 pKa = 4.78YY138 pKa = 10.21TPMEE142 pKa = 4.15VYY144 pKa = 10.89NIVTCDD150 pKa = 2.83ARR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84QLVLSMNARR163 pKa = 11.84LRR165 pKa = 11.84QMHH168 pKa = 7.4GITEE172 pKa = 4.53AFAATILMYY181 pKa = 10.39IATARR186 pKa = 11.84EE187 pKa = 4.18SVAVMIATSSTLWARR202 pKa = 11.84NVDD205 pKa = 3.91LTLLRR210 pKa = 11.84LKK212 pKa = 10.26EE213 pKa = 3.92CSVPFKK219 pKa = 10.6SLHH222 pKa = 5.93HH223 pKa = 7.14ADD225 pKa = 5.26LIDD228 pKa = 3.67LTEE231 pKa = 3.99MFEE234 pKa = 4.76LLSLANRR241 pKa = 11.84GYY243 pKa = 11.2GAVDD247 pKa = 3.0WRR249 pKa = 11.84AEE251 pKa = 4.02KK252 pKa = 10.02EE253 pKa = 4.04HH254 pKa = 6.09RR255 pKa = 11.84TKK257 pKa = 10.83VDD259 pKa = 3.99DD260 pKa = 4.36IKK262 pKa = 11.5VKK264 pKa = 10.83SIDD267 pKa = 3.61VYY269 pKa = 10.74RR270 pKa = 11.84SAMKK274 pKa = 10.13IFEE277 pKa = 4.41MGNKK281 pKa = 9.85NGFRR285 pKa = 11.84YY286 pKa = 9.71KK287 pKa = 10.44KK288 pKa = 10.62LSIKK292 pKa = 10.7DD293 pKa = 3.46FTASRR298 pKa = 11.84WEE300 pKa = 3.82WAPAGSVHH308 pKa = 6.1SQHH311 pKa = 6.62EE312 pKa = 4.18DD313 pKa = 2.94DD314 pKa = 5.55RR315 pKa = 11.84VYY317 pKa = 10.69IDD319 pKa = 3.46PSNYY323 pKa = 7.85RR324 pKa = 11.84HH325 pKa = 5.19RR326 pKa = 11.84TKK328 pKa = 10.6FVTLNSMPARR338 pKa = 11.84HH339 pKa = 6.4IEE341 pKa = 4.13TFLEE345 pKa = 4.28RR346 pKa = 11.84EE347 pKa = 4.35PEE349 pKa = 3.87IQAWPSEE356 pKa = 4.03KK357 pKa = 10.11IEE359 pKa = 3.83WGKK362 pKa = 10.31RR363 pKa = 11.84RR364 pKa = 11.84AIYY367 pKa = 10.16GVDD370 pKa = 3.45LTSATLAHH378 pKa = 6.82LAMYY382 pKa = 10.16NCEE385 pKa = 3.78DD386 pKa = 3.68VFRR389 pKa = 11.84HH390 pKa = 5.87RR391 pKa = 11.84FPVGKK396 pKa = 9.25EE397 pKa = 3.73AEE399 pKa = 3.9ASRR402 pKa = 11.84VHH404 pKa = 5.79KK405 pKa = 10.15RR406 pKa = 11.84LRR408 pKa = 11.84AMCEE412 pKa = 3.92GEE414 pKa = 4.02EE415 pKa = 4.27TFCYY419 pKa = 10.82DD420 pKa = 4.02FDD422 pKa = 5.24DD423 pKa = 5.71FNAQHH428 pKa = 6.05STDD431 pKa = 3.25AMIAVLCAYY440 pKa = 9.48RR441 pKa = 11.84DD442 pKa = 3.51KK443 pKa = 11.38YY444 pKa = 9.67AADD447 pKa = 3.31MSAVQRR453 pKa = 11.84EE454 pKa = 4.19AMDD457 pKa = 3.25WTIASQTKK465 pKa = 9.49TIVNAADD472 pKa = 3.12GRR474 pKa = 11.84YY475 pKa = 7.51RR476 pKa = 11.84TAGTLMSGSRR486 pKa = 11.84LTTFLNTALNFIYY499 pKa = 9.88MDD501 pKa = 3.3IAGVFATPGFRR512 pKa = 11.84DD513 pKa = 3.74SVHH516 pKa = 6.61NGDD519 pKa = 5.1DD520 pKa = 3.32VLLTVEE526 pKa = 4.79SMSTACSIHH535 pKa = 7.26AKK537 pKa = 8.69MAAINARR544 pKa = 11.84AQAAKK549 pKa = 10.35CNAFSIGEE557 pKa = 4.01FLRR560 pKa = 11.84VEE562 pKa = 4.35HH563 pKa = 6.83KK564 pKa = 10.67LSKK567 pKa = 8.94QTGNGAQYY575 pKa = 9.04LTRR578 pKa = 11.84ACATTVHH585 pKa = 6.16SRR587 pKa = 11.84IEE589 pKa = 3.94SQAPVKK595 pKa = 9.12LTSMLGAYY603 pKa = 9.98RR604 pKa = 11.84NRR606 pKa = 11.84QADD609 pKa = 3.16IEE611 pKa = 4.15ARR613 pKa = 11.84AFIPDD618 pKa = 3.62HH619 pKa = 6.87VKK621 pKa = 10.75DD622 pKa = 3.82AMVKK626 pKa = 10.0HH627 pKa = 6.47VINKK631 pKa = 9.09ASTVFTVPKK640 pKa = 8.4EE641 pKa = 4.21TVEE644 pKa = 4.7AIYY647 pKa = 9.83HH648 pKa = 4.89AHH650 pKa = 5.24VVVGGVSTDD659 pKa = 2.89SDD661 pKa = 3.7ASVGVEE667 pKa = 4.19FEE669 pKa = 3.93EE670 pKa = 5.79HH671 pKa = 5.49PAQRR675 pKa = 11.84IFEE678 pKa = 4.6DD679 pKa = 3.62EE680 pKa = 4.26DD681 pKa = 3.78GKK683 pKa = 11.16EE684 pKa = 4.02KK685 pKa = 10.7QLLPGIIDD693 pKa = 3.68YY694 pKa = 11.42SDD696 pKa = 3.93LLASKK701 pKa = 10.67YY702 pKa = 10.97EE703 pKa = 3.96GMLSDD708 pKa = 3.8KK709 pKa = 10.91VVFSAVMRR717 pKa = 11.84ATRR720 pKa = 11.84NQLAVTRR727 pKa = 11.84EE728 pKa = 3.79VEE730 pKa = 3.73IRR732 pKa = 11.84QISTPARR739 pKa = 11.84GKK741 pKa = 10.58FGLARR746 pKa = 11.84SLYY749 pKa = 9.77KK750 pKa = 10.47LYY752 pKa = 11.06KK753 pKa = 10.11NVLQIPFVSQARR765 pKa = 11.84FLGVPPISLATPYY778 pKa = 9.59QAHH781 pKa = 4.84VLKK784 pKa = 10.63RR785 pKa = 11.84VVGNVQDD792 pKa = 3.92PLRR795 pKa = 11.84ALRR798 pKa = 11.84ILLL801 pKa = 4.01
MM1 pKa = 7.35WPVFTRR7 pKa = 11.84MSEE10 pKa = 3.84NRR12 pKa = 11.84LEE14 pKa = 4.06VVDD17 pKa = 3.76RR18 pKa = 11.84HH19 pKa = 5.93KK20 pKa = 10.51CTHH23 pKa = 5.51VLVGVEE29 pKa = 4.44EE30 pKa = 5.03DD31 pKa = 3.53TMTWQGNDD39 pKa = 2.33IMYY42 pKa = 10.43AGQEE46 pKa = 4.29CVRR49 pKa = 11.84TCSYY53 pKa = 11.55GDD55 pKa = 3.31GGTRR59 pKa = 11.84AHH61 pKa = 6.44YY62 pKa = 10.53IKK64 pKa = 10.18MNKK67 pKa = 8.66PVSLLPPIVKK77 pKa = 9.36QRR79 pKa = 11.84LSAAYY84 pKa = 10.12SMVGGYY90 pKa = 10.38VYY92 pKa = 11.01GDD94 pKa = 3.54KK95 pKa = 11.27DD96 pKa = 3.29EE97 pKa = 4.67LASVRR102 pKa = 11.84QILKK106 pKa = 9.66PLKK109 pKa = 9.55QNYY112 pKa = 8.55KK113 pKa = 9.77HH114 pKa = 6.65DD115 pKa = 3.94VKK117 pKa = 11.0FSIRR121 pKa = 11.84SLTYY125 pKa = 9.96KK126 pKa = 10.53KK127 pKa = 10.5SATTGEE133 pKa = 4.3HH134 pKa = 5.44HH135 pKa = 5.79THH137 pKa = 4.78YY138 pKa = 10.21TPMEE142 pKa = 4.15VYY144 pKa = 10.89NIVTCDD150 pKa = 2.83ARR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84QLVLSMNARR163 pKa = 11.84LRR165 pKa = 11.84QMHH168 pKa = 7.4GITEE172 pKa = 4.53AFAATILMYY181 pKa = 10.39IATARR186 pKa = 11.84EE187 pKa = 4.18SVAVMIATSSTLWARR202 pKa = 11.84NVDD205 pKa = 3.91LTLLRR210 pKa = 11.84LKK212 pKa = 10.26EE213 pKa = 3.92CSVPFKK219 pKa = 10.6SLHH222 pKa = 5.93HH223 pKa = 7.14ADD225 pKa = 5.26LIDD228 pKa = 3.67LTEE231 pKa = 3.99MFEE234 pKa = 4.76LLSLANRR241 pKa = 11.84GYY243 pKa = 11.2GAVDD247 pKa = 3.0WRR249 pKa = 11.84AEE251 pKa = 4.02KK252 pKa = 10.02EE253 pKa = 4.04HH254 pKa = 6.09RR255 pKa = 11.84TKK257 pKa = 10.83VDD259 pKa = 3.99DD260 pKa = 4.36IKK262 pKa = 11.5VKK264 pKa = 10.83SIDD267 pKa = 3.61VYY269 pKa = 10.74RR270 pKa = 11.84SAMKK274 pKa = 10.13IFEE277 pKa = 4.41MGNKK281 pKa = 9.85NGFRR285 pKa = 11.84YY286 pKa = 9.71KK287 pKa = 10.44KK288 pKa = 10.62LSIKK292 pKa = 10.7DD293 pKa = 3.46FTASRR298 pKa = 11.84WEE300 pKa = 3.82WAPAGSVHH308 pKa = 6.1SQHH311 pKa = 6.62EE312 pKa = 4.18DD313 pKa = 2.94DD314 pKa = 5.55RR315 pKa = 11.84VYY317 pKa = 10.69IDD319 pKa = 3.46PSNYY323 pKa = 7.85RR324 pKa = 11.84HH325 pKa = 5.19RR326 pKa = 11.84TKK328 pKa = 10.6FVTLNSMPARR338 pKa = 11.84HH339 pKa = 6.4IEE341 pKa = 4.13TFLEE345 pKa = 4.28RR346 pKa = 11.84EE347 pKa = 4.35PEE349 pKa = 3.87IQAWPSEE356 pKa = 4.03KK357 pKa = 10.11IEE359 pKa = 3.83WGKK362 pKa = 10.31RR363 pKa = 11.84RR364 pKa = 11.84AIYY367 pKa = 10.16GVDD370 pKa = 3.45LTSATLAHH378 pKa = 6.82LAMYY382 pKa = 10.16NCEE385 pKa = 3.78DD386 pKa = 3.68VFRR389 pKa = 11.84HH390 pKa = 5.87RR391 pKa = 11.84FPVGKK396 pKa = 9.25EE397 pKa = 3.73AEE399 pKa = 3.9ASRR402 pKa = 11.84VHH404 pKa = 5.79KK405 pKa = 10.15RR406 pKa = 11.84LRR408 pKa = 11.84AMCEE412 pKa = 3.92GEE414 pKa = 4.02EE415 pKa = 4.27TFCYY419 pKa = 10.82DD420 pKa = 4.02FDD422 pKa = 5.24DD423 pKa = 5.71FNAQHH428 pKa = 6.05STDD431 pKa = 3.25AMIAVLCAYY440 pKa = 9.48RR441 pKa = 11.84DD442 pKa = 3.51KK443 pKa = 11.38YY444 pKa = 9.67AADD447 pKa = 3.31MSAVQRR453 pKa = 11.84EE454 pKa = 4.19AMDD457 pKa = 3.25WTIASQTKK465 pKa = 9.49TIVNAADD472 pKa = 3.12GRR474 pKa = 11.84YY475 pKa = 7.51RR476 pKa = 11.84TAGTLMSGSRR486 pKa = 11.84LTTFLNTALNFIYY499 pKa = 9.88MDD501 pKa = 3.3IAGVFATPGFRR512 pKa = 11.84DD513 pKa = 3.74SVHH516 pKa = 6.61NGDD519 pKa = 5.1DD520 pKa = 3.32VLLTVEE526 pKa = 4.79SMSTACSIHH535 pKa = 7.26AKK537 pKa = 8.69MAAINARR544 pKa = 11.84AQAAKK549 pKa = 10.35CNAFSIGEE557 pKa = 4.01FLRR560 pKa = 11.84VEE562 pKa = 4.35HH563 pKa = 6.83KK564 pKa = 10.67LSKK567 pKa = 8.94QTGNGAQYY575 pKa = 9.04LTRR578 pKa = 11.84ACATTVHH585 pKa = 6.16SRR587 pKa = 11.84IEE589 pKa = 3.94SQAPVKK595 pKa = 9.12LTSMLGAYY603 pKa = 9.98RR604 pKa = 11.84NRR606 pKa = 11.84QADD609 pKa = 3.16IEE611 pKa = 4.15ARR613 pKa = 11.84AFIPDD618 pKa = 3.62HH619 pKa = 6.87VKK621 pKa = 10.75DD622 pKa = 3.82AMVKK626 pKa = 10.0HH627 pKa = 6.47VINKK631 pKa = 9.09ASTVFTVPKK640 pKa = 8.4EE641 pKa = 4.21TVEE644 pKa = 4.7AIYY647 pKa = 9.83HH648 pKa = 4.89AHH650 pKa = 5.24VVVGGVSTDD659 pKa = 2.89SDD661 pKa = 3.7ASVGVEE667 pKa = 4.19FEE669 pKa = 3.93EE670 pKa = 5.79HH671 pKa = 5.49PAQRR675 pKa = 11.84IFEE678 pKa = 4.6DD679 pKa = 3.62EE680 pKa = 4.26DD681 pKa = 3.78GKK683 pKa = 11.16EE684 pKa = 4.02KK685 pKa = 10.7QLLPGIIDD693 pKa = 3.68YY694 pKa = 11.42SDD696 pKa = 3.93LLASKK701 pKa = 10.67YY702 pKa = 10.97EE703 pKa = 3.96GMLSDD708 pKa = 3.8KK709 pKa = 10.91VVFSAVMRR717 pKa = 11.84ATRR720 pKa = 11.84NQLAVTRR727 pKa = 11.84EE728 pKa = 3.79VEE730 pKa = 3.73IRR732 pKa = 11.84QISTPARR739 pKa = 11.84GKK741 pKa = 10.58FGLARR746 pKa = 11.84SLYY749 pKa = 9.77KK750 pKa = 10.47LYY752 pKa = 11.06KK753 pKa = 10.11NVLQIPFVSQARR765 pKa = 11.84FLGVPPISLATPYY778 pKa = 9.59QAHH781 pKa = 4.84VLKK784 pKa = 10.63RR785 pKa = 11.84VVGNVQDD792 pKa = 3.92PLRR795 pKa = 11.84ALRR798 pKa = 11.84ILLL801 pKa = 4.01
Molecular weight: 90.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1482 |
681 |
801 |
741.0 |
83.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.232 ± 1.448 | 1.889 ± 0.301 |
5.466 ± 0.021 | 6.005 ± 0.106 |
3.509 ± 0.086 | 5.803 ± 0.623 |
2.767 ± 0.658 | 5.061 ± 0.045 |
5.196 ± 0.421 | 8.907 ± 1.091 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.239 ± 0.198 | 3.981 ± 0.566 |
4.251 ± 0.87 | 3.306 ± 0.143 |
6.478 ± 0.588 | 6.95 ± 0.257 |
5.938 ± 0.427 | 7.895 ± 0.17 |
1.417 ± 0.226 | 3.711 ± 0.122 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
