
Fuerstia marisgermanicae
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Fuerstia
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
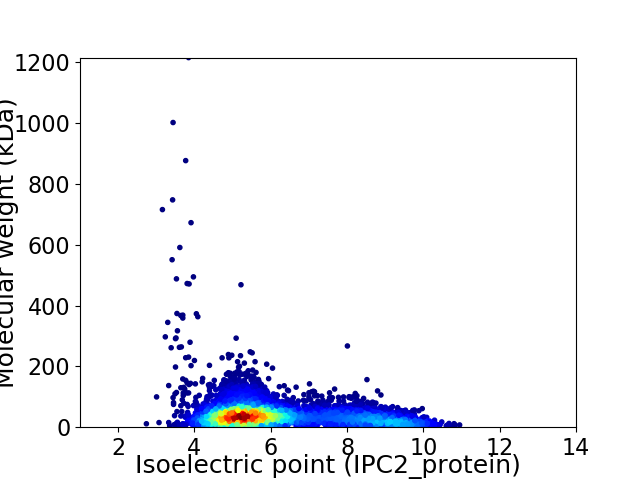
Virtual 2D-PAGE plot for 6466 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8WLJ7|A0A1P8WLJ7_9PLAN Uncharacterized protein OS=Fuerstia marisgermanicae OX=1891926 GN=Fuma_04588 PE=4 SV=1
MM1 pKa = 7.33LQTSNSAGEE10 pKa = 4.11LGAEE14 pKa = 4.37VVLSSLSSIASTKK27 pKa = 10.64LHH29 pKa = 7.31DD30 pKa = 4.23IDD32 pKa = 5.34NDD34 pKa = 3.57GDD36 pKa = 4.22LDD38 pKa = 3.98IVASDD43 pKa = 3.89SGAEE47 pKa = 4.13SILIFINDD55 pKa = 3.55GSANFGAPASVTPDD69 pKa = 3.11EE70 pKa = 4.85TGTTLHH76 pKa = 6.19QVVDD80 pKa = 4.52LNGDD84 pKa = 4.01GYY86 pKa = 11.86ADD88 pKa = 5.03LLVTDD93 pKa = 5.09AGAPNKK99 pKa = 9.74VGYY102 pKa = 9.95YY103 pKa = 8.0PQQAGGTFGARR114 pKa = 11.84VNVPIYY120 pKa = 9.5GQVVSSLSATDD131 pKa = 3.87FNGDD135 pKa = 3.3GVIEE139 pKa = 4.3IGFAHH144 pKa = 6.6TNYY147 pKa = 10.67DD148 pKa = 3.46DD149 pKa = 3.63VAFTFFDD156 pKa = 4.86FNVGWALGQGGGDD169 pKa = 4.11FGPAIVVQRR178 pKa = 11.84YY179 pKa = 9.37DD180 pKa = 3.29SIMYY184 pKa = 10.93SMATPDD190 pKa = 4.2LDD192 pKa = 4.88GDD194 pKa = 4.06GDD196 pKa = 4.22PDD198 pKa = 3.32IVVPQRR204 pKa = 11.84SGPTSVAAFINQDD217 pKa = 3.07GEE219 pKa = 4.17DD220 pKa = 3.9PMRR223 pKa = 11.84FIAPEE228 pKa = 3.71SRR230 pKa = 11.84TYY232 pKa = 10.58TGGDD236 pKa = 3.61PIEE239 pKa = 4.19LQVYY243 pKa = 8.76FGLPITVTGTPRR255 pKa = 11.84IALDD259 pKa = 3.59LGGEE263 pKa = 4.34TVYY266 pKa = 11.22AEE268 pKa = 4.25YY269 pKa = 10.95ASGSGTATHH278 pKa = 5.8QFVYY282 pKa = 10.44QVGEE286 pKa = 4.05TDD288 pKa = 3.79LDD290 pKa = 4.17LDD292 pKa = 4.46GVQLADD298 pKa = 3.64NAIDD302 pKa = 4.1LNEE305 pKa = 4.28GTLTDD310 pKa = 4.15PLGTSALLEE319 pKa = 4.53FPDD322 pKa = 3.97TVLDD326 pKa = 4.03GVVVNAVGPLVQHH339 pKa = 7.03ISRR342 pKa = 11.84GDD344 pKa = 3.62SAPTDD349 pKa = 3.44AASVRR354 pKa = 11.84FTVEE358 pKa = 3.29FSEE361 pKa = 4.37AVEE364 pKa = 4.34GVDD367 pKa = 4.63VSDD370 pKa = 4.53FQVKK374 pKa = 8.48MFDD377 pKa = 3.47GDD379 pKa = 3.64LAGAVIEE386 pKa = 4.62SVTGSGGTYY395 pKa = 9.72EE396 pKa = 4.36VTVSAGTGSGVLALSVNEE414 pKa = 4.17NASIADD420 pKa = 3.86LEE422 pKa = 4.41GAPLARR428 pKa = 11.84GYY430 pKa = 11.04VGGEE434 pKa = 4.08VYY436 pKa = 9.85TLRR439 pKa = 11.84RR440 pKa = 11.84GVEE443 pKa = 3.94APIDD447 pKa = 3.51KK448 pKa = 10.66YY449 pKa = 10.49FTDD452 pKa = 2.9GHH454 pKa = 6.07GDD456 pKa = 3.53YY457 pKa = 10.94RR458 pKa = 11.84PVLNDD463 pKa = 3.58GEE465 pKa = 4.38LSYY468 pKa = 11.31VLHH471 pKa = 7.06GDD473 pKa = 3.34SGVIPDD479 pKa = 4.55GEE481 pKa = 4.46VPSGGIYY488 pKa = 9.88TYY490 pKa = 11.16VDD492 pKa = 2.82STGIVNRR499 pKa = 11.84AEE501 pKa = 4.15DD502 pKa = 3.78DD503 pKa = 3.71SYY505 pKa = 12.24DD506 pKa = 3.84FLGVDD511 pKa = 3.86ANEE514 pKa = 4.1PLYY517 pKa = 10.44VIPSTQVPSLPFLGFSGEE535 pKa = 4.35GVPSGALASYY545 pKa = 10.86VPDD548 pKa = 3.75RR549 pKa = 11.84PNVTSSSASPYY560 pKa = 10.18IKK562 pKa = 10.61LEE564 pKa = 4.05MVDD567 pKa = 3.53VRR569 pKa = 11.84STSGGEE575 pKa = 3.6FSLWSNPPSWNPSAGVTVYY594 pKa = 9.6MATSDD599 pKa = 3.89GVSDD603 pKa = 4.31ADD605 pKa = 3.89ALFLRR610 pKa = 11.84VGSHH614 pKa = 4.51GHH616 pKa = 5.62FNVGFSEE623 pKa = 4.15PGIYY627 pKa = 10.09EE628 pKa = 3.72VDD630 pKa = 3.54IYY632 pKa = 11.68ASGYY636 pKa = 10.24LDD638 pKa = 3.74NDD640 pKa = 3.76HH641 pKa = 6.93NGVYY645 pKa = 10.56DD646 pKa = 4.07PGKK649 pKa = 10.37DD650 pKa = 3.27SYY652 pKa = 11.12IEE654 pKa = 4.31SGIQTMVFAVDD665 pKa = 3.23SLGAVDD671 pKa = 6.02DD672 pKa = 4.38SFTVLEE678 pKa = 4.71GEE680 pKa = 4.46TLTGDD685 pKa = 4.15VSANDD690 pKa = 3.72DD691 pKa = 2.83WHH693 pKa = 6.38EE694 pKa = 4.17AMGEE698 pKa = 4.05YY699 pKa = 8.61TASVEE704 pKa = 4.15TEE706 pKa = 4.07PAHH709 pKa = 6.48GTLTLNADD717 pKa = 3.44GTFTYY722 pKa = 10.5EE723 pKa = 3.76PVAALTGVDD732 pKa = 3.22SFAYY736 pKa = 10.32RR737 pKa = 11.84LTNEE741 pKa = 4.01RR742 pKa = 11.84GGFTTATVTLLTHH755 pKa = 6.71HH756 pKa = 6.83SPTAVEE762 pKa = 4.17DD763 pKa = 4.06AFVVASGSSVYY774 pKa = 11.11GNVLFNDD781 pKa = 3.93FDD783 pKa = 4.37VDD785 pKa = 4.06GDD787 pKa = 4.18ALLAALDD794 pKa = 4.34AGPTSGSVVVSADD807 pKa = 2.83GSFVYY812 pKa = 10.17TPSAAFDD819 pKa = 4.32DD820 pKa = 4.58SDD822 pKa = 4.06SFTYY826 pKa = 10.68SIDD829 pKa = 3.85DD830 pKa = 3.81GTGLASTATVVISAAEE846 pKa = 3.87DD847 pKa = 3.6AEE849 pKa = 4.46FEE851 pKa = 4.51VVLTEE856 pKa = 3.7GHH858 pKa = 5.63VDD860 pKa = 2.81IGVVIGEE867 pKa = 4.39HH868 pKa = 6.75DD869 pKa = 3.79HH870 pKa = 8.13DD871 pKa = 4.85EE872 pKa = 5.59DD873 pKa = 5.71EE874 pKa = 5.95DD875 pKa = 4.48DD876 pKa = 5.77HH877 pKa = 9.71DD878 pKa = 6.91DD879 pKa = 5.8DD880 pKa = 6.59DD881 pKa = 5.3DD882 pKa = 5.26HH883 pKa = 8.94DD884 pKa = 4.84HH885 pKa = 6.87EE886 pKa = 5.98EE887 pKa = 3.8EE888 pKa = 4.68AEE890 pKa = 4.08WDD892 pKa = 3.79LHH894 pKa = 5.27VHH896 pKa = 4.91DD897 pKa = 5.39HH898 pKa = 6.08EE899 pKa = 6.24GEE901 pKa = 4.32VEE903 pKa = 4.08YY904 pKa = 10.64HH905 pKa = 6.73ADD907 pKa = 3.47EE908 pKa = 4.66ALLYY912 pKa = 10.91VGMDD916 pKa = 4.81AITDD920 pKa = 4.04RR921 pKa = 11.84PAGTAYY927 pKa = 10.74DD928 pKa = 4.88FIGTDD933 pKa = 2.92AGEE936 pKa = 4.52SVFVLPATEE945 pKa = 4.4NSDD948 pKa = 4.62LLFLGLGTEE957 pKa = 4.35EE958 pKa = 4.53IEE960 pKa = 4.61VGTFLDD966 pKa = 3.95GTLEE970 pKa = 3.96LRR972 pKa = 11.84LKK974 pKa = 10.49SVSGPGQFSMWTSGLEE990 pKa = 4.14GPEE993 pKa = 3.79VAMATSDD1000 pKa = 4.21GITEE1004 pKa = 4.52ADD1006 pKa = 4.84LITLLEE1012 pKa = 4.73AGHH1015 pKa = 6.26AHH1017 pKa = 6.62ANFGFTEE1024 pKa = 3.96MGLYY1028 pKa = 10.28QITLQASGTLADD1040 pKa = 4.24GDD1042 pKa = 3.81VSEE1045 pKa = 4.91DD1046 pKa = 3.01VTYY1049 pKa = 10.39FFKK1052 pKa = 11.09VGNTAEE1058 pKa = 4.74GIEE1061 pKa = 4.32VQNGMTQRR1069 pKa = 11.84SYY1071 pKa = 11.17IDD1073 pKa = 3.49EE1074 pKa = 4.91LDD1076 pKa = 3.66VLFGSEE1082 pKa = 5.5DD1083 pKa = 3.64GLDD1086 pKa = 4.82DD1087 pKa = 4.64ILTNPDD1093 pKa = 2.66RR1094 pKa = 11.84VQITKK1099 pKa = 10.52YY1100 pKa = 10.68DD1101 pKa = 3.86LNGEE1105 pKa = 4.21NGSLLSDD1112 pKa = 3.61AGYY1115 pKa = 10.69SLSANGSRR1123 pKa = 11.84LSFNFGVQGLGGNRR1137 pKa = 11.84NSNAGDD1143 pKa = 3.84GYY1145 pKa = 10.94YY1146 pKa = 10.29EE1147 pKa = 4.17IGIDD1151 pKa = 3.47ADD1153 pKa = 3.84GDD1155 pKa = 4.23GEE1157 pKa = 4.55YY1158 pKa = 9.11EE1159 pKa = 4.46TYY1161 pKa = 10.59QSFYY1165 pKa = 11.27RR1166 pKa = 11.84LLGDD1170 pKa = 3.31VNGDD1174 pKa = 3.68RR1175 pKa = 11.84KK1176 pKa = 10.53VDD1178 pKa = 3.24AADD1181 pKa = 3.76RR1182 pKa = 11.84MSVMQSMRR1190 pKa = 11.84NQNPEE1195 pKa = 3.59ADD1197 pKa = 3.8VNGDD1201 pKa = 3.19GRR1203 pKa = 11.84VNSIDD1208 pKa = 3.5FVLVSRR1214 pKa = 11.84ALGRR1218 pKa = 11.84DD1219 pKa = 3.26LRR1221 pKa = 11.84DD1222 pKa = 3.24GLFDD1226 pKa = 5.63DD1227 pKa = 6.27DD1228 pKa = 4.63EE1229 pKa = 6.9DD1230 pKa = 4.0EE1231 pKa = 4.25
MM1 pKa = 7.33LQTSNSAGEE10 pKa = 4.11LGAEE14 pKa = 4.37VVLSSLSSIASTKK27 pKa = 10.64LHH29 pKa = 7.31DD30 pKa = 4.23IDD32 pKa = 5.34NDD34 pKa = 3.57GDD36 pKa = 4.22LDD38 pKa = 3.98IVASDD43 pKa = 3.89SGAEE47 pKa = 4.13SILIFINDD55 pKa = 3.55GSANFGAPASVTPDD69 pKa = 3.11EE70 pKa = 4.85TGTTLHH76 pKa = 6.19QVVDD80 pKa = 4.52LNGDD84 pKa = 4.01GYY86 pKa = 11.86ADD88 pKa = 5.03LLVTDD93 pKa = 5.09AGAPNKK99 pKa = 9.74VGYY102 pKa = 9.95YY103 pKa = 8.0PQQAGGTFGARR114 pKa = 11.84VNVPIYY120 pKa = 9.5GQVVSSLSATDD131 pKa = 3.87FNGDD135 pKa = 3.3GVIEE139 pKa = 4.3IGFAHH144 pKa = 6.6TNYY147 pKa = 10.67DD148 pKa = 3.46DD149 pKa = 3.63VAFTFFDD156 pKa = 4.86FNVGWALGQGGGDD169 pKa = 4.11FGPAIVVQRR178 pKa = 11.84YY179 pKa = 9.37DD180 pKa = 3.29SIMYY184 pKa = 10.93SMATPDD190 pKa = 4.2LDD192 pKa = 4.88GDD194 pKa = 4.06GDD196 pKa = 4.22PDD198 pKa = 3.32IVVPQRR204 pKa = 11.84SGPTSVAAFINQDD217 pKa = 3.07GEE219 pKa = 4.17DD220 pKa = 3.9PMRR223 pKa = 11.84FIAPEE228 pKa = 3.71SRR230 pKa = 11.84TYY232 pKa = 10.58TGGDD236 pKa = 3.61PIEE239 pKa = 4.19LQVYY243 pKa = 8.76FGLPITVTGTPRR255 pKa = 11.84IALDD259 pKa = 3.59LGGEE263 pKa = 4.34TVYY266 pKa = 11.22AEE268 pKa = 4.25YY269 pKa = 10.95ASGSGTATHH278 pKa = 5.8QFVYY282 pKa = 10.44QVGEE286 pKa = 4.05TDD288 pKa = 3.79LDD290 pKa = 4.17LDD292 pKa = 4.46GVQLADD298 pKa = 3.64NAIDD302 pKa = 4.1LNEE305 pKa = 4.28GTLTDD310 pKa = 4.15PLGTSALLEE319 pKa = 4.53FPDD322 pKa = 3.97TVLDD326 pKa = 4.03GVVVNAVGPLVQHH339 pKa = 7.03ISRR342 pKa = 11.84GDD344 pKa = 3.62SAPTDD349 pKa = 3.44AASVRR354 pKa = 11.84FTVEE358 pKa = 3.29FSEE361 pKa = 4.37AVEE364 pKa = 4.34GVDD367 pKa = 4.63VSDD370 pKa = 4.53FQVKK374 pKa = 8.48MFDD377 pKa = 3.47GDD379 pKa = 3.64LAGAVIEE386 pKa = 4.62SVTGSGGTYY395 pKa = 9.72EE396 pKa = 4.36VTVSAGTGSGVLALSVNEE414 pKa = 4.17NASIADD420 pKa = 3.86LEE422 pKa = 4.41GAPLARR428 pKa = 11.84GYY430 pKa = 11.04VGGEE434 pKa = 4.08VYY436 pKa = 9.85TLRR439 pKa = 11.84RR440 pKa = 11.84GVEE443 pKa = 3.94APIDD447 pKa = 3.51KK448 pKa = 10.66YY449 pKa = 10.49FTDD452 pKa = 2.9GHH454 pKa = 6.07GDD456 pKa = 3.53YY457 pKa = 10.94RR458 pKa = 11.84PVLNDD463 pKa = 3.58GEE465 pKa = 4.38LSYY468 pKa = 11.31VLHH471 pKa = 7.06GDD473 pKa = 3.34SGVIPDD479 pKa = 4.55GEE481 pKa = 4.46VPSGGIYY488 pKa = 9.88TYY490 pKa = 11.16VDD492 pKa = 2.82STGIVNRR499 pKa = 11.84AEE501 pKa = 4.15DD502 pKa = 3.78DD503 pKa = 3.71SYY505 pKa = 12.24DD506 pKa = 3.84FLGVDD511 pKa = 3.86ANEE514 pKa = 4.1PLYY517 pKa = 10.44VIPSTQVPSLPFLGFSGEE535 pKa = 4.35GVPSGALASYY545 pKa = 10.86VPDD548 pKa = 3.75RR549 pKa = 11.84PNVTSSSASPYY560 pKa = 10.18IKK562 pKa = 10.61LEE564 pKa = 4.05MVDD567 pKa = 3.53VRR569 pKa = 11.84STSGGEE575 pKa = 3.6FSLWSNPPSWNPSAGVTVYY594 pKa = 9.6MATSDD599 pKa = 3.89GVSDD603 pKa = 4.31ADD605 pKa = 3.89ALFLRR610 pKa = 11.84VGSHH614 pKa = 4.51GHH616 pKa = 5.62FNVGFSEE623 pKa = 4.15PGIYY627 pKa = 10.09EE628 pKa = 3.72VDD630 pKa = 3.54IYY632 pKa = 11.68ASGYY636 pKa = 10.24LDD638 pKa = 3.74NDD640 pKa = 3.76HH641 pKa = 6.93NGVYY645 pKa = 10.56DD646 pKa = 4.07PGKK649 pKa = 10.37DD650 pKa = 3.27SYY652 pKa = 11.12IEE654 pKa = 4.31SGIQTMVFAVDD665 pKa = 3.23SLGAVDD671 pKa = 6.02DD672 pKa = 4.38SFTVLEE678 pKa = 4.71GEE680 pKa = 4.46TLTGDD685 pKa = 4.15VSANDD690 pKa = 3.72DD691 pKa = 2.83WHH693 pKa = 6.38EE694 pKa = 4.17AMGEE698 pKa = 4.05YY699 pKa = 8.61TASVEE704 pKa = 4.15TEE706 pKa = 4.07PAHH709 pKa = 6.48GTLTLNADD717 pKa = 3.44GTFTYY722 pKa = 10.5EE723 pKa = 3.76PVAALTGVDD732 pKa = 3.22SFAYY736 pKa = 10.32RR737 pKa = 11.84LTNEE741 pKa = 4.01RR742 pKa = 11.84GGFTTATVTLLTHH755 pKa = 6.71HH756 pKa = 6.83SPTAVEE762 pKa = 4.17DD763 pKa = 4.06AFVVASGSSVYY774 pKa = 11.11GNVLFNDD781 pKa = 3.93FDD783 pKa = 4.37VDD785 pKa = 4.06GDD787 pKa = 4.18ALLAALDD794 pKa = 4.34AGPTSGSVVVSADD807 pKa = 2.83GSFVYY812 pKa = 10.17TPSAAFDD819 pKa = 4.32DD820 pKa = 4.58SDD822 pKa = 4.06SFTYY826 pKa = 10.68SIDD829 pKa = 3.85DD830 pKa = 3.81GTGLASTATVVISAAEE846 pKa = 3.87DD847 pKa = 3.6AEE849 pKa = 4.46FEE851 pKa = 4.51VVLTEE856 pKa = 3.7GHH858 pKa = 5.63VDD860 pKa = 2.81IGVVIGEE867 pKa = 4.39HH868 pKa = 6.75DD869 pKa = 3.79HH870 pKa = 8.13DD871 pKa = 4.85EE872 pKa = 5.59DD873 pKa = 5.71EE874 pKa = 5.95DD875 pKa = 4.48DD876 pKa = 5.77HH877 pKa = 9.71DD878 pKa = 6.91DD879 pKa = 5.8DD880 pKa = 6.59DD881 pKa = 5.3DD882 pKa = 5.26HH883 pKa = 8.94DD884 pKa = 4.84HH885 pKa = 6.87EE886 pKa = 5.98EE887 pKa = 3.8EE888 pKa = 4.68AEE890 pKa = 4.08WDD892 pKa = 3.79LHH894 pKa = 5.27VHH896 pKa = 4.91DD897 pKa = 5.39HH898 pKa = 6.08EE899 pKa = 6.24GEE901 pKa = 4.32VEE903 pKa = 4.08YY904 pKa = 10.64HH905 pKa = 6.73ADD907 pKa = 3.47EE908 pKa = 4.66ALLYY912 pKa = 10.91VGMDD916 pKa = 4.81AITDD920 pKa = 4.04RR921 pKa = 11.84PAGTAYY927 pKa = 10.74DD928 pKa = 4.88FIGTDD933 pKa = 2.92AGEE936 pKa = 4.52SVFVLPATEE945 pKa = 4.4NSDD948 pKa = 4.62LLFLGLGTEE957 pKa = 4.35EE958 pKa = 4.53IEE960 pKa = 4.61VGTFLDD966 pKa = 3.95GTLEE970 pKa = 3.96LRR972 pKa = 11.84LKK974 pKa = 10.49SVSGPGQFSMWTSGLEE990 pKa = 4.14GPEE993 pKa = 3.79VAMATSDD1000 pKa = 4.21GITEE1004 pKa = 4.52ADD1006 pKa = 4.84LITLLEE1012 pKa = 4.73AGHH1015 pKa = 6.26AHH1017 pKa = 6.62ANFGFTEE1024 pKa = 3.96MGLYY1028 pKa = 10.28QITLQASGTLADD1040 pKa = 4.24GDD1042 pKa = 3.81VSEE1045 pKa = 4.91DD1046 pKa = 3.01VTYY1049 pKa = 10.39FFKK1052 pKa = 11.09VGNTAEE1058 pKa = 4.74GIEE1061 pKa = 4.32VQNGMTQRR1069 pKa = 11.84SYY1071 pKa = 11.17IDD1073 pKa = 3.49EE1074 pKa = 4.91LDD1076 pKa = 3.66VLFGSEE1082 pKa = 5.5DD1083 pKa = 3.64GLDD1086 pKa = 4.82DD1087 pKa = 4.64ILTNPDD1093 pKa = 2.66RR1094 pKa = 11.84VQITKK1099 pKa = 10.52YY1100 pKa = 10.68DD1101 pKa = 3.86LNGEE1105 pKa = 4.21NGSLLSDD1112 pKa = 3.61AGYY1115 pKa = 10.69SLSANGSRR1123 pKa = 11.84LSFNFGVQGLGGNRR1137 pKa = 11.84NSNAGDD1143 pKa = 3.84GYY1145 pKa = 10.94YY1146 pKa = 10.29EE1147 pKa = 4.17IGIDD1151 pKa = 3.47ADD1153 pKa = 3.84GDD1155 pKa = 4.23GEE1157 pKa = 4.55YY1158 pKa = 9.11EE1159 pKa = 4.46TYY1161 pKa = 10.59QSFYY1165 pKa = 11.27RR1166 pKa = 11.84LLGDD1170 pKa = 3.31VNGDD1174 pKa = 3.68RR1175 pKa = 11.84KK1176 pKa = 10.53VDD1178 pKa = 3.24AADD1181 pKa = 3.76RR1182 pKa = 11.84MSVMQSMRR1190 pKa = 11.84NQNPEE1195 pKa = 3.59ADD1197 pKa = 3.8VNGDD1201 pKa = 3.19GRR1203 pKa = 11.84VNSIDD1208 pKa = 3.5FVLVSRR1214 pKa = 11.84ALGRR1218 pKa = 11.84DD1219 pKa = 3.26LRR1221 pKa = 11.84DD1222 pKa = 3.24GLFDD1226 pKa = 5.63DD1227 pKa = 6.27DD1228 pKa = 4.63EE1229 pKa = 6.9DD1230 pKa = 4.0EE1231 pKa = 4.25
Molecular weight: 129.78 kDa
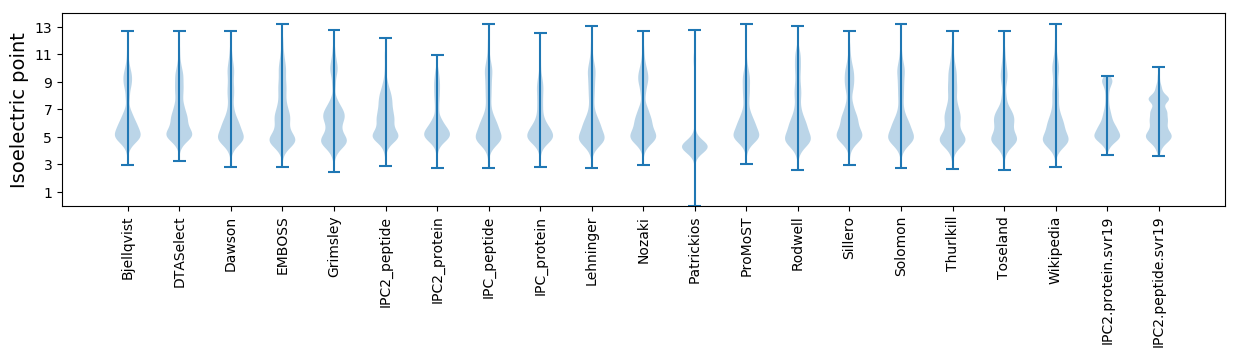
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8WL87|A0A1P8WL87_9PLAN Uncharacterized protein OS=Fuerstia marisgermanicae OX=1891926 GN=Fuma_04454 PE=4 SV=1
MM1 pKa = 7.1QRR3 pKa = 11.84AKK5 pKa = 10.82LNPPPKK11 pKa = 9.25TNLVSLLISTFIGAVTGAVVLAPFTRR37 pKa = 11.84SFGAPTGRR45 pKa = 11.84GVGFGLGGMISMIGALIVRR64 pKa = 11.84GLGRR68 pKa = 11.84RR69 pKa = 11.84LTLL72 pKa = 3.73
MM1 pKa = 7.1QRR3 pKa = 11.84AKK5 pKa = 10.82LNPPPKK11 pKa = 9.25TNLVSLLISTFIGAVTGAVVLAPFTRR37 pKa = 11.84SFGAPTGRR45 pKa = 11.84GVGFGLGGMISMIGALIVRR64 pKa = 11.84GLGRR68 pKa = 11.84RR69 pKa = 11.84LTLL72 pKa = 3.73
Molecular weight: 7.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2532177 |
30 |
12079 |
391.6 |
43.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.809 ± 0.034 | 1.15 ± 0.018 |
6.501 ± 0.044 | 5.894 ± 0.04 |
3.811 ± 0.017 | 7.61 ± 0.059 |
2.241 ± 0.021 | 4.881 ± 0.028 |
3.782 ± 0.047 | 9.409 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.182 ± 0.021 | 3.55 ± 0.036 |
5.001 ± 0.029 | 4.035 ± 0.028 |
6.278 ± 0.053 | 6.63 ± 0.034 |
6.115 ± 0.077 | 7.387 ± 0.028 |
1.466 ± 0.015 | 2.267 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
